Ontology application and use at the ENCODE DCC
- PMID: 25776021
- PMCID: PMC4360730
- DOI: 10.1093/database/bav010
Ontology application and use at the ENCODE DCC
Abstract
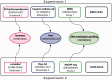
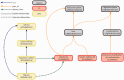
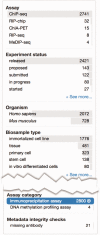
The Encyclopedia of DNA elements (ENCODE) project is an ongoing collaborative effort to create a catalog of genomic annotations. To date, the project has generated over 4000 experiments across more than 350 cell lines and tissues using a wide array of experimental techniques to study the chromatin structure, regulatory network and transcriptional landscape of the Homo sapiens and Mus musculus genomes. All ENCODE experimental data, metadata and associated computational analyses are submitted to the ENCODE Data Coordination Center (DCC) for validation, tracking, storage and distribution to community resources and the scientific community. As the volume of data increases, the organization of experimental details becomes increasingly complicated and demands careful curation to identify related experiments. Here, we describe the ENCODE DCC's use of ontologies to standardize experimental metadata. We discuss how ontologies, when used to annotate metadata, provide improved searching capabilities and facilitate the ability to find connections within a set of experiments. Additionally, we provide examples of how ontologies are used to annotate ENCODE metadata and how the annotations can be identified via ontology-driven searches at the ENCODE portal. As genomic datasets grow larger and more interconnected, standardization of metadata becomes increasingly vital to allow for exploration and comparison of data between different scientific projects.
© The Author(s) 2015. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

