DNA methylation and hydroxymethylation in stem cells
- PMID: 25776144
- PMCID: PMC4687961
- DOI: 10.1002/cbf.3101
DNA methylation and hydroxymethylation in stem cells
Abstract
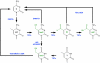
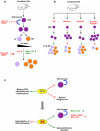
In mammals, DNA methylation and hydroxymethylation are specific epigenetic mechanisms that can contribute to the regulation of gene expression and cellular functions. DNA methylation is important for the function of embryonic stem cells and adult stem cells (such as haematopoietic stem cells, neural stem cells and germline stem cells), and changes in DNA methylation patterns are essential for successful nuclear reprogramming. In the past several years, the rediscovery of hydroxymethylation and the TET enzymes expanded our insights tremendously and uncovered more dynamic aspects of cytosine methylation regulation. Here, we review the current knowledge and highlight the most recent advances in DNA methylation and hydroxymethylation in embryonic stem cells, induced pluripotent stem cells and several well-studied adult stems cells. Our current understanding of stem cell epigenetics and new advances in the field will undoubtedly stimulate further clinical applications of regenerative medicine in the future.
Keywords: adult stem cells; epigenetics; hydroxymethylation; methylation; stem cells.
Copyright © 2015 John Wiley & Sons, Ltd.
Figures
References
-
- Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170:1072–1073. doi:10.1038/1701072a0. - PubMed
-
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi:10.1016/j.cell.2007.11.019. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi:10.1016/j.cell.2006.07.024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

