The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity
- PMID: 25776556
- PMCID: PMC4405639
- DOI: 10.1128/MCB.01521-14
The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity
Abstract
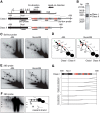
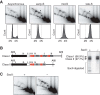
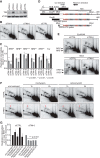
In S phase, the replication and transcription of genomic DNA need to accommodate each other, otherwise their machineries collide, with chromosomal instability as a possible consequence. Here, we characterized the human replication fork barrier (RFB) that is present downstream from the 47S pre-rRNA gene (ribosomal DNA [rDNA]). We found that the most proximal transcription terminator, Sal box T1, acts as a polar RFB, while the other, Sal box T4/T5, arrests replication forks bidirectionally. The fork-arresting activity at these sites depends on polymerase I (Pol I) transcription termination factor 1 (TTF-1) and a replisome component, TIMELESS (TIM). We also found that the RFB activity was linked to rDNA copies with hypomethylated CpG and coincided with the time that actively transcribed rRNA genes are replicated. Failed fork arrest at RFB sites led to a slowdown of fork progression moving in the opposite direction to rRNA transcription. Chemical inhibition of transcription counteracted this deceleration of forks, indicating that rRNA transcription impedes replication in the absence of RFB activity. Thus, our results reveal a role of RFB for coordinating the progression of replication and transcription activity in highly transcribed rRNA genes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Dalgaard JZ, Godfrey EL, MacFarlane RJ. 2011. Eukaryotic replication barriers: how, why and where forks stall, chap 13 In Seligmann H. (ed), DNA replication: current advances. InTech, Rijeka, Croatia. doi:10.5772/20383. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
