Quo vadis? Microbial profiling revealed strong effects of cleanroom maintenance and routes of contamination in indoor environments
- PMID: 25778463
- PMCID: PMC4361859
- DOI: 10.1038/srep09156
Quo vadis? Microbial profiling revealed strong effects of cleanroom maintenance and routes of contamination in indoor environments
Abstract
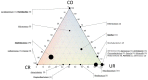
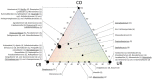
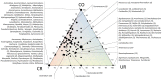
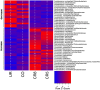
Space agencies maintain highly controlled cleanrooms to ensure the demands of planetary protection. To study potential effects of microbiome control, we analyzed microbial communities in two particulate-controlled cleanrooms (ISO 5 and ISO 8) and two vicinal uncontrolled areas (office, changing room) by cultivation and 16S rRNA gene amplicon analysis (cloning, pyrotagsequencing, and PhyloChip G3 analysis). Maintenance procedures affected the microbiome on total abundance and microbial community structure concerning richness, diversity and relative abundance of certain taxa. Cleanroom areas were found to be mainly predominated by potentially human-associated bacteria; archaeal signatures were detected in every area. Results indicate that microorganisms were mainly spread from the changing room (68%) into the cleanrooms, potentially carried along with human activity. The numbers of colony forming units were reduced by up to ~400 fold from the uncontrolled areas towards the ISO 5 cleanroom, accompanied with a reduction of the living portion of microorganisms from 45% (changing area) to 1% of total 16S rRNA gene signatures as revealed via propidium monoazide treatment of the samples. Our results demonstrate the strong effects of cleanroom maintenance on microbial communities in indoor environments and can be used to improve the design and operation of biologically controlled cleanrooms.
Figures

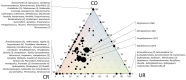
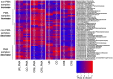
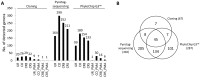
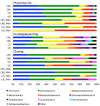

References
-
- Gilbert J. A. & Dupont C. L. Microbial metagenomics: beyond the genome. Ann Rev Mar Sci 3, 347–371 (2011). - PubMed
-
- Nocker A., Cheung C.-Y. & Camper A. K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67, 310–320 (2006). - PubMed
-
- Huber T., Faulkner G. & Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

