HIV-1 integrase multimerization as a therapeutic target
- PMID: 25778682
- PMCID: PMC4791179
- DOI: 10.1007/82_2015_439
HIV-1 integrase multimerization as a therapeutic target
Abstract
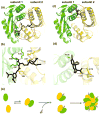
Multimeric HIV-1 integrase (IN) plays an essential, multifunctional role in virus replication and serves as an important therapeutic target. Structural and biochemical studies have revealed the importance of the ordered interplay between IN molecules for its function. In the presence of viral DNA ends, individual IN subunits assemble into a tetramer and form a stable synaptic complex (SSC), which mediates integration of the reverse transcribed HIV-1 genome into chromatin. Cellular chromatin-associated protein LEDGF/p75 engages the IN tetramer in the SSC and directs HIV-1 integration into active genes. A mechanism to deregulate the productive interplay between IN subunits with small molecule inhibitors has recently received considerable attention. Most notably, allosteric IN inhibitors (ALLINIs) have been shown to bind to the IN dimer interface at the LEDGF/p75 binding pocket, stabilize interacting IN subunits, and promote aberrant, higher order IN multimerization. Consequently, these compounds impair formation of the SSC and associated LEDGF/p75-independent IN catalytic activities as well as inhibit LEDGF/p75 binding to the SSC in vitro. However, in infected cells, ALLINIs more potently impaired correct maturation of virus particles than the integration step. ALLINI treatments induced aberrant, higher order IN multimerization in virions and resulted in eccentric, non-infectious virus particles. These studies have suggested that the correctly ordered IN structure is important for virus particle morphogenesis and highlighted IN multimerization as a plausible therapeutic target for developing new inhibitors to enhance treatment options for HIV-1-infected patients.
Figures
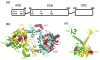
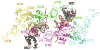
References
-
- Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PloS One. 2013;8(9):e74163. doi: 10.1371/journal.pone.0074163. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

