The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis
- PMID: 25778917
- PMCID: PMC4362455
- DOI: 10.1083/jcb.201410047
The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis
Abstract
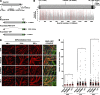
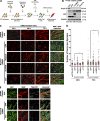
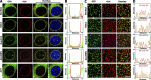
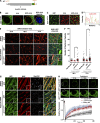
Previously, we identified the nucleoporin gp210/Nup210 as a critical regulator of muscle and neuronal differentiation, but how this nucleoporin exerts its function and whether it modulates nuclear pore complex (NPC) activity remain unknown. Here, we show that gp210/Nup210 mediates muscle cell differentiation in vitro via its conserved N-terminal domain that extends into the perinuclear space. Removal of the C-terminal domain, which partially mislocalizes gp210/Nup210 away from NPCs, efficiently rescues the differentiation defect caused by the knockdown of endogenous gp210/Nup210. Unexpectedly, a gp210/Nup210 mutant lacking the NPC-targeting transmembrane and C-terminal domains is sufficient for C2C12 myoblast differentiation. We demonstrate that the endoplasmic reticulum (ER) stress-specific caspase cascade is exacerbated during Nup210 depletion and that blocking ER stress-mediated apoptosis rescues differentiation of Nup210-deficient cells. Our results suggest that the role of gp210/Nup210 in cell differentiation is mediated by its large luminal domain, which can act independently of NPC association and appears to play a pivotal role in the maintenance of nuclear envelope/ER homeostasis.
© 2015 Gomez-Cavazos and Hetzer.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

