VaRank: a simple and powerful tool for ranking genetic variants
- PMID: 25780760
- PMCID: PMC4358652
- DOI: 10.7717/peerj.796
VaRank: a simple and powerful tool for ranking genetic variants
Abstract
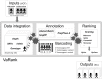
Background. Most genetic disorders are caused by single nucleotide variations (SNVs) or small insertion/deletions (indels). High throughput sequencing has broadened the catalogue of human variation, including common polymorphisms, rare variations or disease causing mutations. However, identifying one variation among hundreds or thousands of others is still a complex task for biologists, geneticists and clinicians. Results. We have developed VaRank, a command-line tool for the ranking of genetic variants detected by high-throughput sequencing. VaRank scores and prioritizes variants annotated either by Alamut Batch or SnpEff. A barcode allows users to quickly view the presence/absence of variants (with homozygote/heterozygote status) in analyzed samples. VaRank supports the commonly used VCF input format for variants analysis thus allowing it to be easily integrated into NGS bioinformatics analysis pipelines. VaRank has been successfully applied to disease-gene identification as well as to molecular diagnostics setup for several hundred patients. Conclusions. VaRank is implemented in Tcl/Tk, a scripting language which is platform-independent but has been tested only on Unix environment. The source code is available under the GNU GPL, and together with sample data and detailed documentation can be downloaded from http://www.lbgi.fr/VaRank/.
Keywords: Annotation; Barcode; Human genetics; Molecular diagnostic; Mutation detection; Next generation sequencing; Software; Variant ranking.
Conflict of interest statement
Co-author André Blavier declares financial competing interest as a member of Interactive Biosoftware, the company commercializing Alamut Batch.
Figures




References
-
- Aldahmesh MA, Li Y, Alhashem A, Anazi S, Alkuraya H, Hashem M, Awaji AA, Sogaty S, Alkharashi A, Alzahrani S, Al Hazzaa SA, Xiong Y, Kong S, Sun Z, Alkuraya FS. IFT27, encoding a small GTPase component of IFT particles, is mutated in a consanguineous family with Bardet-Biedl syndrome. Human Molecular Genetics. 2014;23:3307–3315. doi: 10.1093/hmg/ddu044. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

