Genetic interaction between Tmprss2-ERG gene fusion and Nkx3.1-loss does not enhance prostate tumorigenesis in mouse models
- PMID: 25780911
- PMCID: PMC4364018
- DOI: 10.1371/journal.pone.0120628
Genetic interaction between Tmprss2-ERG gene fusion and Nkx3.1-loss does not enhance prostate tumorigenesis in mouse models
Abstract
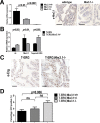
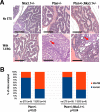
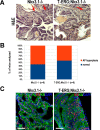
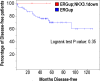
Gene fusions involving ETS family transcription factors (mainly TMPRSS2-ERG and TMPRSS2-ETV1 fusions) have been found in ~50% of human prostate cancer cases. Although expression of TMPRSS2-ERG or TMPRSS2-ETV1 fusion alone is insufficient to initiate prostate tumorigenesis, they appear to sensitize prostate epithelial cells for cooperation with additional oncogenic mutations to drive frank prostate adenocarcinoma. To search for such ETS-cooperating oncogenic events, we focused on a well-studied prostate tumor suppressor NKX3.1, as loss of NKX3.1 is another common genetic alteration in human prostate cancer. Previous studies have shown that deletions at 8p21 (harboring NKX3.1) and 21q22 (resulting in TMPRSS2-ERG fusion) were both present in a subtype of prostate cancer cases, and that ERG can lead to epigenetic silencing of NKX3.1 in prostate cancer cells, whereas NKX3.1 can in turn negatively regulate TMPRSS2-ERG fusion expression via suppression of the TMPRSS2 promoter activity. We recently generated knockin mouse models for TMPRSS2-ERG and TMPRSS2-ETV1 fusions, utilizing the endogenous Tmprss2 promoter. We crossed these knockin models to an Nkx3.1 knockout mouse model. In Tmprss2-ERG;Nkx3.1+/- (or -/-) male mice, although we observed a slight but significant upregulation of Tmprss2-ERG fusion expression upon Nkx3.1 loss, we did not detect any significant cooperation between these two genetic events to enhance prostate tumorigenesis in vivo. Furthermore, retrospective analysis of a previously published human prostate cancer dataset revealed that within ERG-overexpressing prostate cancer cases, NKX3.1 loss or deletion did not predict biochemical relapse after radical prostatectomy. Collectively, these data suggest that although TMPRSS2-ERG fusion and loss of NKX3.1 are among the most common mutational events found in prostate cancer, and although each of them can sensitize prostate epithelial cells for cooperating with other oncogenic events, these two events themselves do not appear to cooperate at a significant level in vivo to enhance prostate tumorigenesis.
Conflict of interest statement
Figures
Similar articles
-
Loss of the NKX3.1 tumorsuppressor promotes the TMPRSS2-ERG fusion gene expression in prostate cancer.BMC Cancer. 2014 Jan 13;14:16. doi: 10.1186/1471-2407-14-16. BMC Cancer. 2014. PMID: 24418414 Free PMC article.
-
Binding of TMPRSS2-ERG to BAF Chromatin Remodeling Complexes Mediates Prostate Oncogenesis.Mol Cell. 2018 Aug 16;71(4):554-566.e7. doi: 10.1016/j.molcel.2018.06.040. Epub 2018 Aug 2. Mol Cell. 2018. PMID: 30078722 Free PMC article.
-
TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer.Cancer Res. 2006 Nov 15;66(22):10658-63. doi: 10.1158/0008-5472.CAN-06-1871. Cancer Res. 2006. PMID: 17108102
-
[The progress of TMPRSS2-ETS gene fusions and their mechanism in prostate cancer].Yi Chuan. 2011 Feb;33(2):117-22. doi: 10.3724/sp.j.1005.2011.00117. Yi Chuan. 2011. PMID: 21377967 Review. Chinese.
-
Prevalence and clinical application of TMPRSS2-ERG fusion in Asian prostate cancer patients: a large-sample study in Chinese people and a systematic review.Asian J Androl. 2020 Mar-Apr;22(2):200-207. doi: 10.4103/aja.aja_45_19. Asian J Androl. 2020. PMID: 31210145 Free PMC article.
Cited by
-
Genetics and biology of prostate cancer.Genes Dev. 2018 Sep 1;32(17-18):1105-1140. doi: 10.1101/gad.315739.118. Genes Dev. 2018. PMID: 30181359 Free PMC article. Review.
-
Function and regulation of the PEA3 subfamily of ETS transcription factors in cancer.Am J Cancer Res. 2020 Oct 1;10(10):3083-3105. eCollection 2020. Am J Cancer Res. 2020. PMID: 33163259 Free PMC article. Review.
-
Genetically Engineered Mouse Models of Prostate Cancer in the Postgenomic Era.Cold Spring Harb Perspect Med. 2019 Feb 1;9(2):a030528. doi: 10.1101/cshperspect.a030528. Cold Spring Harb Perspect Med. 2019. PMID: 29661807 Free PMC article. Review.
References
-
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310: 644–648. - PubMed
-
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448: 595–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

