Crystal and EM structures of human phosphoribosyl pyrophosphate synthase I (PRS1) provide novel insights into the disease-associated mutations
- PMID: 25781187
- PMCID: PMC4363470
- DOI: 10.1371/journal.pone.0120304
Crystal and EM structures of human phosphoribosyl pyrophosphate synthase I (PRS1) provide novel insights into the disease-associated mutations
Abstract
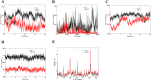
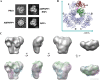
Human PRS1, which is indispensable for the biosynthesis of nucleotides, deoxynucleotides and their derivatives, is associated directly with multiple human diseases because of single base mutation. However, a molecular understanding of the effect of these mutations is hampered by the lack of understanding of its catalytic mechanism. Here, we reconstruct the 3D EM structure of the PRS1 apo state. Together with the native stain EM structures of AMPNPP, AMPNPP and R5P, ADP and the apo states with distinct conformations, we suggest the hexamer is the enzymatically active form. Based on crystal structures, sequence analysis, mutagenesis, enzyme kinetics assays, and MD simulations, we reveal the conserved substrates binding motifs and make further analysis of all pathogenic mutants.
Conflict of interest statement
Figures
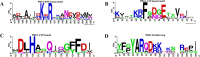
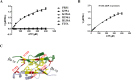


Similar articles
-
A small disturbance, but a serious disease: the possible mechanism of D52H-mutant of human PRS1 that causes gout.IUBMB Life. 2013 Jun;65(6):518-25. doi: 10.1002/iub.1154. Epub 2013 Mar 18. IUBMB Life. 2013. PMID: 23509005
-
Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site.Biochem J. 2007 Jan 1;401(1):39-47. doi: 10.1042/BJ20061066. Biochem J. 2007. PMID: 16939420 Free PMC article.
-
Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.Nat Struct Biol. 2000 Apr;7(4):303-8. doi: 10.1038/74069. Nat Struct Biol. 2000. PMID: 10742175
-
Phosphoribosylpyrophosphate synthetase and the regulation of phosphoribosylpyrophosphate production in human cells.Prog Nucleic Acid Res Mol Biol. 2001;69:115-48. doi: 10.1016/s0079-6603(01)69046-9. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550793 Review.
-
[Structure of family proteins constituting mammalian phosphoribosylpyrophosphate synthetase].Seikagaku. 1997 Feb;69(2):122-6. Seikagaku. 1997. PMID: 9086843 Review. Japanese. No abstract available.
Cited by
-
Conversion of PRPS Hexamer to Monomer by AMPK-Mediated Phosphorylation Inhibits Nucleotide Synthesis in Response to Energy Stress.Cancer Discov. 2018 Jan;8(1):94-107. doi: 10.1158/2159-8290.CD-17-0712. Epub 2017 Oct 26. Cancer Discov. 2018. PMID: 29074724 Free PMC article.
-
Molecular mechanism of c-Myc and PRPS1/2 against thiopurine resistance in Burkitt's lymphoma.J Cell Mol Med. 2020 Jun;24(12):6704-6715. doi: 10.1111/jcmm.15322. Epub 2020 May 11. J Cell Mol Med. 2020. PMID: 32391636 Free PMC article.
-
The role of gene duplication and paralog specialisation in the evolution of the mammalian PRPS complex.Nat Commun. 2025 Jul 8;16(1):6076. doi: 10.1038/s41467-025-61216-z. Nat Commun. 2025. PMID: 40628709 Free PMC article.
-
Human PRPS1 filaments stabilize allosteric sites to regulate activity.Nat Struct Mol Biol. 2023 Mar;30(3):391-402. doi: 10.1038/s41594-023-00921-z. Epub 2023 Feb 6. Nat Struct Mol Biol. 2023. PMID: 36747094 Free PMC article.
-
Functional characterization of a novel loss-of-function mutation of PRPS1 related to early-onset progressive nonsyndromic hearing loss in Koreans (DFNX1): Potential implications on future therapeutic intervention.J Gene Med. 2016 Nov;18(11-12):353-358. doi: 10.1002/jgm.2935. J Gene Med. 2016. PMID: 27886419 Free PMC article.
References
-
- Kamal MA, Christopherson RI (2004) Accumulation of 5-phosphoribosyl-1-pyrophosphate in human CCRF-CEM leukaemia cells treated with antifolates. Int J Biochem Cell Biol 36: 545–551. - PubMed
-
- Fisher DI, Safrany ST, Strike P, McLennan AG, Cartwright JL (2002) Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J Biol Chem 277: 47313–47317. - PubMed
-
- Kadziola A, Jepsen CH, Johansson E, McGuire J, Larsen S, Hove-Jensen B (2005) Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J Mol Biol 354: 815–828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

