The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in Fusarium graminearum
- PMID: 25781642
- PMCID: PMC6638501
- DOI: 10.1111/mpp.12254
The AreA transcription factor mediates the regulation of deoxynivalenol (DON) synthesis by ammonium and cyclic adenosine monophosphate (cAMP) signalling in Fusarium graminearum
Abstract
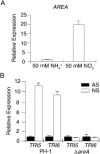
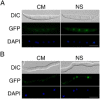
Deoxynivalenol (DON), a trichothecene mycotoxin produced by Fusarium graminearum, is harmful to humans and animals. Because different nitrogen sources are known to have opposite effects on DON production, in this study, we characterized the regulatory mechanisms of the AREA transcription factor in trichothecene biosynthesis. The ΔareA mutant showed significantly reduced vegetative growth and DON production in cultures inoculated with hyphae. Suppression of TRI gene expression and DON production by ammonium were diminished in the ΔareA mutant. The deletion of AREA also affected the stimulatory effects of arginine on DON biosynthesis. The AreA-green fluorescent protein (GFP) fusion complemented the ΔareA mutant, and its localization to the nucleus was enhanced under nitrogen starvation conditions. Site-directed mutagenesis showed that the conserved predicted protein kinase A (PKA) phosphorylation site S874 was important for AreA function, indicating that AreA may be a downstream target of the cyclic adenosine monophosphate (cAMP)-PKA pathway, which is known to regulate DON production. We also showed that AreA interacted with Tri10 in co-immunoprecipitation assays. The interaction of AreA with Tri10 is probably related to its role in the regulation of TRI gene expression. Interestingly, the ΔareA mutant showed significantly reduced PKA activity and expression of all three predicted ammonium permease (MEP) genes, in particular MEP1, under low ammonium conditions. Taken together, our results show that AREA is involved in the regulation of DON production by ammonium suppression and the cAMP-PKA pathway. The AreA transcription factor may interact with Tri10 and control the expression and up-regulation of MEP genes.
Keywords: DON production; Gibberella zeae; TRI6 expression; ammonium suppression; nitrogen metabolism.
© 2015 BSPP AND JOHN WILEY & SONS LTD.
Figures




Similar articles
-
MeJA inhibits fungal growth and DON toxin production by interfering with the cAMP-PKA signaling pathway in the wheat scab fungus Fusarium graminearum.mBio. 2025 Mar 12;16(3):e0315124. doi: 10.1128/mbio.03151-24. Epub 2025 Feb 4. mBio. 2025. PMID: 39902906 Free PMC article.
-
TRI6 and TRI10 play different roles in the regulation of deoxynivalenol (DON) production by cAMP signalling in Fusarium graminearum.Environ Microbiol. 2016 Nov;18(11):3689-3701. doi: 10.1111/1462-2920.13279. Epub 2016 Apr 21. Environ Microbiol. 2016. PMID: 26940955
-
The cyclase-associated protein FgCap1 has both protein kinase A-dependent and -independent functions during deoxynivalenol production and plant infection in Fusarium graminearum.Mol Plant Pathol. 2018 Mar;19(3):552-563. doi: 10.1111/mpp.12540. Epub 2017 Mar 23. Mol Plant Pathol. 2018. PMID: 28142217 Free PMC article.
-
Toxic mechanisms of the trichothecenes T-2 toxin and deoxynivalenol on protein synthesis.Food Chem Toxicol. 2022 Jun;164:113044. doi: 10.1016/j.fct.2022.113044. Epub 2022 Apr 19. Food Chem Toxicol. 2022. PMID: 35452771 Review.
-
Towards systems biology of mycotoxin regulation.Toxins (Basel). 2013 Apr 18;5(4):675-82. doi: 10.3390/toxins5040675. Toxins (Basel). 2013. PMID: 23598563 Free PMC article. Review.
Cited by
-
Transcriptome analysis reveals the regulatory mode by which NAA promotes the growth of Armillaria gallica.PLoS One. 2022 Nov 21;17(11):e0277701. doi: 10.1371/journal.pone.0277701. eCollection 2022. PLoS One. 2022. PMID: 36409681 Free PMC article.
-
MeJA inhibits fungal growth and DON toxin production by interfering with the cAMP-PKA signaling pathway in the wheat scab fungus Fusarium graminearum.mBio. 2025 Mar 12;16(3):e0315124. doi: 10.1128/mbio.03151-24. Epub 2025 Feb 4. mBio. 2025. PMID: 39902906 Free PMC article.
-
Comparative Transcriptomic Analyses Propose the Molecular Regulatory Mechanisms Underlying 1,8-Cineole from Cinnamomum kanehirae Hay and Promote the Asexual Sporulation of Antrodia cinnamomea in Submerged Fermentation.Molecules. 2023 Nov 9;28(22):7511. doi: 10.3390/molecules28227511. Molecules. 2023. PMID: 38005233 Free PMC article. Review.
-
Combination of Pseudo-LC-NMR and HRMS/MS-Based Molecular Networking for the Rapid Identification of Antimicrobial Metabolites From Fusarium petroliphilum.Front Mol Biosci. 2021 Oct 22;8:725691. doi: 10.3389/fmolb.2021.725691. eCollection 2021. Front Mol Biosci. 2021. PMID: 34746230 Free PMC article.
-
GATA-type transcriptional factor SpGAT1 interacts with SpMIG1 and promotes lipid accumulation in the oleaginous yeast Saitozyma podzolica zwy-2-3.Biotechnol Biofuels Bioprod. 2022 Oct 8;15(1):103. doi: 10.1186/s13068-022-02177-z. Biotechnol Biofuels Bioprod. 2022. PMID: 36209175 Free PMC article.
References
-
- Alexander, N.J. , Proctor, R.H. and McCormick, S.P. (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 28, 198–215.
-
- Bai, G.H. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. - PubMed
-
- Beck, T. and Hall, M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient‐regulated transcription factors. Nature, 402, 689–692. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

