Molecular dynamics simulation and experimental verification of the interaction between cyclin T1 and HIV-1 Tat proteins
- PMID: 25781978
- PMCID: PMC4363469
- DOI: 10.1371/journal.pone.0119451
Molecular dynamics simulation and experimental verification of the interaction between cyclin T1 and HIV-1 Tat proteins
Abstract
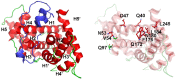
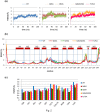
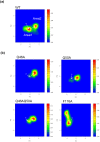
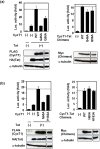
The viral encoded Tat protein is essential for the transcriptional activation of HIV proviral DNA. Interaction of Tat with a cellular transcription elongation factor P-TEFb containing CycT1 is critically required for its action. In this study, we performed MD simulation using the 3D data for wild-type and 4CycT1mutants3D data. We found that the dynamic structural change of CycT1 H2' helix is indispensable for its activity for the Tat action. Moreover, we detected flexible structural changes of the Tat-recognition cavity in the WT CycT1 comprising of ten AAs that are in contact with Tat. These structural fluctuations in WT were lost in the CycT1 mutants. We also found the critical importance of the hydrogen bond network involving H1, H1' and H2 helices of CycT1. Since similar AA substitutions of the Tat-CycT1 chimera retained the Tat-supporting activity, these interactions are considered primarily involved in interaction with Tat. These findings described in this paper should provide vital information for the development of effective anti-Tat compound.
Conflict of interest statement
Figures

Similar articles
-
MD simulation of the Tat/Cyclin T1/CDK9 complex revealing the hidden catalytic cavity within the CDK9 molecule upon Tat binding.PLoS One. 2017 Feb 8;12(2):e0171727. doi: 10.1371/journal.pone.0171727. eCollection 2017. PLoS One. 2017. PMID: 28178316 Free PMC article.
-
Functional characterization of human cyclin T1 N-terminal region for human immunodeficiency virus-1 Tat transcriptional activation.J Mol Biol. 2011 Jul 29;410(5):887-95. doi: 10.1016/j.jmb.2011.04.061. J Mol Biol. 2011. PMID: 21763494
-
Cyclin T1 stabilizes expression levels of HIV-1 Tat in cells.FEBS J. 2009 Dec;276(23):7124-33. doi: 10.1111/j.1742-4658.2009.07424.x. FEBS J. 2009. PMID: 20064163
-
Fab'-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography.J Mol Biol. 2011 Jan 7;405(1):33-42. doi: 10.1016/j.jmb.2010.10.033. Epub 2010 Oct 28. J Mol Biol. 2011. PMID: 21035463 Review.
-
Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression.J Cell Biochem. 1999 Dec 1;75(3):357-68. J Cell Biochem. 1999. PMID: 10536359 Review.
Cited by
-
Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: A computational approach.PLoS One. 2017 Apr 14;12(4):e0174953. doi: 10.1371/journal.pone.0174953. eCollection 2017. PLoS One. 2017. PMID: 28410371 Free PMC article.
-
Ensemble-based modeling and rigidity decomposition of allosteric interaction networks and communication pathways in cyclin-dependent kinases: Differentiating kinase clients of the Hsp90-Cdc37 chaperone.PLoS One. 2017 Nov 2;12(11):e0186089. doi: 10.1371/journal.pone.0186089. eCollection 2017. PLoS One. 2017. PMID: 29095844 Free PMC article.
-
HIV Tat/P-TEFb Interaction: A Potential Target for Novel Anti-HIV Therapies.Molecules. 2018 Apr 17;23(4):933. doi: 10.3390/molecules23040933. Molecules. 2018. PMID: 29673219 Free PMC article. Review.
-
Perspectives towards antiviral drug discovery against Ebola virus.J Med Virol. 2019 Dec;91(12):2029-2048. doi: 10.1002/jmv.25357. Epub 2019 Sep 30. J Med Virol. 2019. PMID: 30431654 Free PMC article. Review.
-
MD simulation of the Tat/Cyclin T1/CDK9 complex revealing the hidden catalytic cavity within the CDK9 molecule upon Tat binding.PLoS One. 2017 Feb 8;12(2):e0171727. doi: 10.1371/journal.pone.0171727. eCollection 2017. PLoS One. 2017. PMID: 28178316 Free PMC article.
References
-
- Joint United Nations Program on HIV/AIDS (UNAIDS). Grobal report: UNAIDS report on the global AIDS epidemic 2013. http://www.unaids.org.
-
- Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350: 1023–1035. - PubMed
-
- Karn J. Tackling Tat. J Mol Biol. 1999;293: 235–254. - PubMed
-
- Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin BM. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264: 245–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

