Comparison of locus-specific databases for BRCA1 and BRCA2 variants reveals disparity in variant classification within and among databases
- PMID: 25782689
- PMCID: PMC4567983
- DOI: 10.1007/s12687-015-0220-x
Comparison of locus-specific databases for BRCA1 and BRCA2 variants reveals disparity in variant classification within and among databases
Abstract
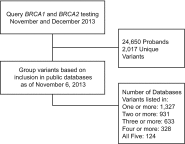
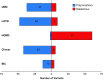
Genetic variants of uncertain clinical significance (VUSs) are a common outcome of clinical genetic testing. Locus-specific variant databases (LSDBs) have been established for numerous disease-associated genes as a research tool for the interpretation of genetic sequence variants to facilitate variant interpretation via aggregated data. If LSDBs are to be used for clinical practice, consistent and transparent criteria regarding the deposition and interpretation of variants are vital, as variant classifications are often used to make important and irreversible clinical decisions. In this study, we performed a retrospective analysis of 2017 consecutive BRCA1 and BRCA2 genetic variants identified from 24,650 consecutive patient samples referred to our laboratory to establish an unbiased dataset representative of the types of variants seen in the US patient population, submitted by clinicians and researchers for BRCA1 and BRCA2 testing. We compared the clinical classifications of these variants among five publicly accessible BRCA1 and BRCA2 variant databases: BIC, ClinVar, HGMD (paid version), LOVD, and the UMD databases. Our results show substantial disparity of variant classifications among publicly accessible databases. Furthermore, it appears that discrepant classifications are not the result of a single outlier but widespread disagreement among databases. This study also shows that databases sometimes favor a clinical classification when current best practice guidelines (ACMG/AMP/CAP) would suggest an uncertain classification. Although LSDBs have been well established for research applications, our results suggest several challenges preclude their wider use in clinical practice.
Keywords: BRCA1; BRCA2; Locus-specific variant databases; Variant classification; Variants of uncertain significance.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

