Outside the coding genome, mammalian microRNAs confer structural and functional complexity
- PMID: 25783159
- PMCID: PMC4425368
- DOI: 10.1126/scisignal.2005813
Outside the coding genome, mammalian microRNAs confer structural and functional complexity
Abstract
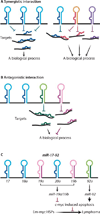
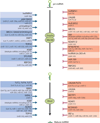
MicroRNAs (miRNAs) comprise a class of small, regulatory noncoding RNAs (ncRNAs) with pivotal roles in posttranscriptional gene regulation. Since their initial discovery in 1993, numerous miRNAs have been identified in mammalian genomes, many of which play important roles in diverse cellular processes in development and disease. These small ncRNAs regulate the expression of many protein-coding genes posttranscriptionally, thus adding a substantial complexity to the molecular networks underlying physiological development and disease. In part, this complexity arises from the distinct gene structures, the extensive genomic redundancy, and the complex regulation of the expression and biogenesis of miRNAs. These characteristics contribute to the functional robustness and versatility of miRNAs and provide important clues to the functional significance of these small ncRNAs. The unique structure and function of miRNAs will continue to inspire many to explore the vast noncoding genome and to elucidate the molecular basis for the functional complexity of mammalian genomes.
Copyright © 2015, American Association for the Advancement of Science.
Figures
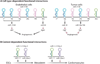


References
-
- Mattick JS. Legislative Activity: RNA regulation: a new genetics? 2004;5:1662–1666.
-
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

