The Arabidopsis RNA-binding protein AtRGGA regulates tolerance to salt and drought stress
- PMID: 25783413
- PMCID: PMC4424017
- DOI: 10.1104/pp.114.255802
The Arabidopsis RNA-binding protein AtRGGA regulates tolerance to salt and drought stress
Abstract
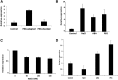
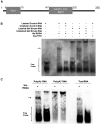
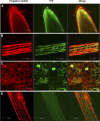
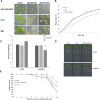
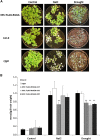
Salt and drought stress severely reduce plant growth and crop productivity worldwide. The identification of genes underlying stress response and tolerance is the subject of intense research in plant biology. Through microarray analyses, we previously identified in potato (Solanum tuberosum) StRGGA, coding for an Arginine Glycine Glycine (RGG) box-containing RNA-binding protein, whose expression was specifically induced in potato cell cultures gradually exposed to osmotic stress. Here, we show that the Arabidopsis (Arabidopsis thaliana) ortholog, AtRGGA, is a functional RNA-binding protein required for a proper response to osmotic stress. AtRGGA gene expression was up-regulated in seedlings after long-term exposure to abscisic acid (ABA) and polyethylene glycol, while treatments with NaCl resulted in AtRGGA down-regulation. AtRGGA promoter analysis showed activity in several tissues, including stomata, the organs controlling transpiration. Fusion of AtRGGA with yellow fluorescent protein indicated that AtRGGA is localized in the cytoplasm and the cytoplasmic perinuclear region. In addition, the rgga knockout mutant was hypersensitive to ABA in root growth and survival tests and to salt stress during germination and at the vegetative stage. AtRGGA-overexpressing plants showed higher tolerance to ABA and salt stress on plates and in soil, accumulating lower levels of proline when exposed to drought stress. Finally, a global analysis of gene expression revealed extensive alterations in the transcriptome under salt stress, including several genes such as ASCORBATE PEROXIDASE2, GLUTATHIONE S-TRANSFERASE TAU9, and several SMALL AUXIN UPREGULATED RNA-like genes showing opposite expression behavior in transgenic and knockout plants. Taken together, our results reveal an important role of AtRGGA in the mechanisms of plant response and adaptation to stress.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Albà MM, Pagès M (1998) Plant proteins containing the RNA-recognition motif. Trends Plant Sci 3: 15–21
-
- Ambrosone A, Costa A, Leone A, Grillo S (2012) Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci 182: 12–18 - PubMed
-
- Ambrosone A, Costa A, Martinelli R, Massarelli I, De Simone V, Grillo S, Leone A (2011) Differential gene regulation in potato cells and plants upon abrupt or gradual exposure to water stress. Acta Physiol Plant 33: 1157–1171
-
- Ambrosone A, Di Giacomo M, Leone A, Grillo MS, Costa A (2013) Identification of early induced genes upon water deficit in potato cell cultures by cDNA-AFLP. J Plant Res 126: 169–178 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

