Bacterial actin and tubulin homologs in cell growth and division
- PMID: 25784047
- PMCID: PMC5519336
- DOI: 10.1016/j.cub.2015.01.030
Bacterial actin and tubulin homologs in cell growth and division
Abstract
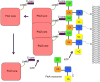
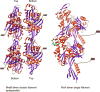
In contrast to the elaborate cytoskeletal machines harbored by eukaryotic cells, such as mitotic spindles, cytoskeletal structures detectable by typical negative stain electron microscopy are generally absent from bacterial cells. As a result, for decades it was thought that bacteria lacked cytoskeletal machines. Revolutions in genomics and fluorescence microscopy have confirmed the existence not only of smaller-scale cytoskeletal structures in bacteria, but also of widespread functional homologs of eukaryotic cytoskeletal proteins. The presence of actin, tubulin, and intermediate filament homologs in these relatively simple cells suggests that primitive cytoskeletons first arose in bacteria. In bacteria such as Escherichia coli, homologs of tubulin and actin directly interact with each other and are crucial for coordinating cell growth and division. The function and direct interactions between these proteins will be the focus of this review.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


References
-
- Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. - PubMed
-
- Gitai Z. Diversification and specialization of the bacterial cytoskeleton. Curr Opin Cell Biol. 2007;19:5–12. - PubMed
-
- Pogliano J. The bacterial cytoskeleton. Curr Opin Cell Biol. 2008;20:19–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

