Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles
- PMID: 25784292
- PMCID: PMC4503522
- DOI: 10.1002/ijc.29517
Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles
Abstract
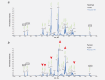
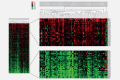
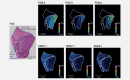
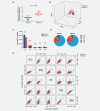
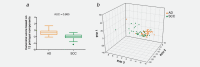
Non-small cell lung cancer (NSCLC) is the leading cause of cancer death globally. To develop better diagnostics and more effective treatments, research in the past decades has focused on identification of molecular changes in the genome, transcriptome, proteome, and more recently also the metabolome. Phospholipids, which nevertheless play a central role in cell functioning, remain poorly explored. Here, using a mass spectrometry (MS)-based phospholipidomics approach, we profiled 179 phospholipid species in malignant and matched non-malignant lung tissue of 162 NSCLC patients (73 in a discovery cohort and 89 in a validation cohort). We identified 91 phospholipid species that were differentially expressed in cancer versus non-malignant tissues. Most prominent changes included a decrease in sphingomyelins (SMs) and an increase in specific phosphatidylinositols (PIs). Also a decrease in multiple phosphatidylserines (PSs) was observed, along with an increase in several phosphatidylethanolamine (PE) and phosphatidylcholine (PC) species, particularly those with 40 or 42 carbon atoms in both fatty acyl chains together. 2D-imaging MS of the most differentially expressed phospholipids confirmed their differential abundance in cancer cells. We identified lipid markers that can discriminate tumor versus normal tissue and different NSCLC subtypes with an AUC (area under the ROC curve) of 0.999 and 0.885, respectively. In conclusion, using both shotgun and 2D-imaging lipidomics analysis, we uncovered a hitherto unrecognized alteration in phospholipid profiles in NSCLC. These changes may have important biological implications and may have significant potential for biomarker development.
Keywords: 2D-imaging MS; lipidomics; mass spectrometry; non-small cell lung cancer; phospholipids.
© 2015 The Authors. Published by Wiley Periodicals, Inc. on behalf of UICC.
Figures
References
-
- Globocan; Available at. In: Cancer IAfRo, ed., 2012. http://globocan.iarc.fr/. (Accessed on: Sept 30, 2014)
-
- Tan DS, Camilleri-Broet S, Tan EH. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. 2014;135:1092–100. , et al. Int J Cancer. - PubMed
-
- Zhang L, Zhang W, Chen K. Search for cancer risk factors with microarray-based genome-wide association studies. Technol Cancer Res Treat. 2010;9:107–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

