Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis
- PMID: 25784702
- PMCID: PMC4453511
- DOI: 10.1128/mBio.02428-14
Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis
Abstract
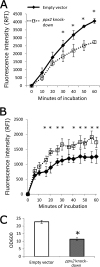
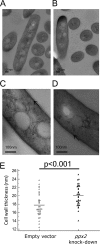
Mycobacterium tuberculosis can persist for decades in the human host. Stringent response pathways involving inorganic polyphosphate [poly(P)], which is synthesized and hydrolyzed by polyphosphate kinase (PPK) and exopolyphosphatase (PPX), respectively, are believed to play a key regulatory role in bacterial persistence. We show here that M. tuberculosis poly(P) accumulation is temporally linked to bacillary growth restriction. We also identify M. tuberculosis Rv1026 as a novel exopolyphosphatase with hydrolytic activity against long-chain poly(P). Using a tetracycline-inducible expression system to knock down expression of Rv1026 (ppx2), we found that M. tuberculosis poly(P) accumulation leads to slowed growth and reduced susceptibility to isoniazid, increased resistance to heat and acid pH, and enhanced intracellular survival during macrophage infection. By transmission electron microscopy, the ppx2 knockdown strain exhibited increased cell wall thickness, which was associated with reduced cell wall permeability to hydrophilic drugs rather than induction of drug efflux pumps or altered biofilm formation relative to the empty vector control. Transcriptomic and metabolomic analysis revealed a metabolic downshift of the ppx2 knockdown characterized by reduced transcription and translation and a downshift of glycerol-3-phosphate levels. In summary, poly(P) plays an important role in M. tuberculosis growth restriction and metabolic downshift and contributes to antibiotic tolerance through altered cell wall permeability.
Importance: The stringent response, involving the regulatory molecules inorganic polyphosphate [poly(P)] and (p)ppGpp, is believed to mediate Mycobacterium tuberculosis persistence. In this study, we identified a novel enzyme (Rv1026, PPX2) responsible for hydrolyzing long-chain poly(P). A genetically engineered M. tuberculosis strain deficient in the ppx2 gene showed increased poly(P) levels, which were associated with early bacterial growth arrest and reduced susceptibility to the first-line drug isoniazid, as well as increased bacterial survival during exposure to stress conditions and within macrophages. Relative to the control strain, the mutant showed increased thickness of the cell wall and reduced drug permeability. Global gene expression and metabolite analysis revealed reduced expression of the transcriptional and translational machinery and a shift in carbon source utilization. In summary, regulation of the poly(P) balance is critical for persister formation in M. tuberculosis.
Copyright © 2015 Chuang et al.
Figures

Similar articles
-
Stringent Response Factors PPX1 and PPK2 Play an Important Role in Mycobacterium tuberculosis Metabolism, Biofilm Formation, and Sensitivity to Isoniazid In Vivo.Antimicrob Agents Chemother. 2016 Oct 21;60(11):6460-6470. doi: 10.1128/AAC.01139-16. Print 2016 Nov. Antimicrob Agents Chemother. 2016. PMID: 27527086 Free PMC article.
-
The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence.PLoS One. 2011;6(11):e28076. doi: 10.1371/journal.pone.0028076. Epub 2011 Nov 21. PLoS One. 2011. PMID: 22132215 Free PMC article.
-
The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence.mBio. 2013 May 21;4(3):e00039-13. doi: 10.1128/mBio.00039-13. mBio. 2013. PMID: 23695835 Free PMC article.
-
[Polyphosphate and its physiological function in Mycobacteria - A review].Wei Sheng Wu Xue Bao. 2016 Dec 4;56(12):1840-6. Wei Sheng Wu Xue Bao. 2016. PMID: 29741848 Review. Chinese.
-
[Progress in polyphosphate and related metabolizing enzymes].Sheng Li Ke Xue Jin Zhan. 2011 Jun;42(3):181-7. Sheng Li Ke Xue Jin Zhan. 2011. PMID: 21932515 Review. Chinese.
Cited by
-
Proteomic and Transcriptomic Analyses Indicate Reduced Biofilm-Forming Abilities in Cefiderocol-Resistant Klebsiella pneumoniae.Front Microbiol. 2022 Jan 3;12:778190. doi: 10.3389/fmicb.2021.778190. eCollection 2021. Front Microbiol. 2022. PMID: 35046911 Free PMC article.
-
Purification, crystallization and X-ray crystallographic analysis of a putative exopolyphosphatase from Zymomonas mobilis.Acta Crystallogr F Struct Biol Commun. 2016 Mar;72(Pt 3):172-8. doi: 10.1107/S2053230X16000753. Epub 2016 Feb 19. Acta Crystallogr F Struct Biol Commun. 2016. PMID: 26919520 Free PMC article.
-
Insights on recent approaches in drug discovery strategies and untapped drug targets against drug resistance.Futur J Pharm Sci. 2021;7(1):56. doi: 10.1186/s43094-021-00196-5. Epub 2021 Mar 3. Futur J Pharm Sci. 2021. PMID: 33686369 Free PMC article. Review.
-
Hypoxic Non-replicating Persistent Mycobacterium tuberculosis Develops Thickened Outer Layer That Helps in Restricting Rifampicin Entry.Front Microbiol. 2019 Oct 11;10:2339. doi: 10.3389/fmicb.2019.02339. eCollection 2019. Front Microbiol. 2019. PMID: 31681204 Free PMC article.
-
Stringent Response in Mycobacteria: From Biology to Therapeutic Potential.Pathogens. 2021 Nov 1;10(11):1417. doi: 10.3390/pathogens10111417. Pathogens. 2021. PMID: 34832573 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

