Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment
- PMID: 25784703
- PMCID: PMC4453514
- DOI: 10.1128/mBio.02471-14
Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment
Abstract
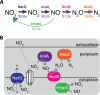
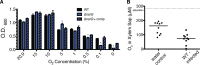
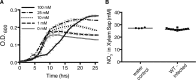
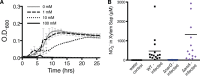
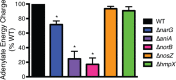
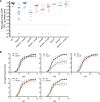
Genomic data predict that, in addition to oxygen, the bacterial plant pathogen Ralstonia solanacearum can use nitrate (NO3(-)), nitrite (NO2(-)), nitric oxide (NO), and nitrous oxide (N2O) as terminal electron acceptors (TEAs). Genes encoding inorganic nitrogen reduction were highly expressed during tomato bacterial wilt disease, when the pathogen grows in xylem vessels. Direct measurements found that tomato xylem fluid was low in oxygen, especially in plants infected by R. solanacearum. Xylem fluid contained ~25 mM NO3(-), corresponding to R. solanacearum's optimal NO3(-) concentration for anaerobic growth in vitro. We tested the hypothesis that R. solanacearum uses inorganic nitrogen species to respire and grow during pathogenesis by making deletion mutants that each lacked a step in nitrate respiration (ΔnarG), denitrification (ΔaniA, ΔnorB, and ΔnosZ), or NO detoxification (ΔhmpX). The ΔnarG, ΔaniA, and ΔnorB mutants grew poorly on NO3(-) compared to the wild type, and they had reduced adenylate energy charge levels under anaerobiosis. While NarG-dependent NO3(-) respiration directly enhanced growth, AniA-dependent NO2(-) reduction did not. NO2(-) and NO inhibited growth in culture, and their removal depended on denitrification and NO detoxification. Thus, NO3(-) acts as a TEA, but the resulting NO2(-) and NO likely do not. None of the mutants grew as well as the wild type in planta, and strains lacking AniA (NO2(-) reductase) or HmpX (NO detoxification) had reduced virulence on tomato. Thus, R. solanacearum exploits host NO3(-) to respire, grow, and cause disease. Degradation of NO2(-) and NO is also important for successful infection and depends on denitrification and NO detoxification systems.
Importance: The plant-pathogenic bacterium Ralstonia solanacearum causes bacterial wilt, one of the world's most destructive crop diseases. This pathogen's explosive growth in plant vascular xylem is poorly understood. We used biochemical and genetic approaches to show that R. solanacearum rapidly depletes oxygen in host xylem but can then respire using host nitrate as a terminal electron acceptor. The microbe uses its denitrification pathway to detoxify the reactive nitrogen species nitrite (a product of nitrate respiration) and nitric oxide (a plant defense signal). Detoxification may play synergistic roles in bacterial wilt virulence by converting the host's chemical weapon into an energy source. Mutant bacterial strains lacking elements of the denitrification pathway could not grow as well as the wild type in tomato plants, and some mutants were also reduced in virulence. Our results show how a pathogen's metabolic activity can alter the host environment in ways that increase pathogen success.
Copyright © 2015 Dalsing et al.
Figures

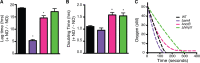
Similar articles
-
Plant-Pathogenic Ralstonia Phylotypes Evolved Divergent Respiratory Strategies and Behaviors To Thrive in Xylem.mBio. 2023 Feb 28;14(1):e0318822. doi: 10.1128/mbio.03188-22. Epub 2023 Feb 6. mBio. 2023. PMID: 36744950 Free PMC article.
-
NorA, HmpX, and NorB Cooperate to Reduce NO Toxicity during Denitrification and Plant Pathogenesis in Ralstonia solanacearum.Microbiol Spectr. 2022 Apr 27;10(2):e0026422. doi: 10.1128/spectrum.00264-22. Epub 2022 Apr 4. Microbiol Spectr. 2022. PMID: 35377234 Free PMC article.
-
The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato.mBio. 2012 Aug 31;3(4):e00114-12. doi: 10.1128/mBio.00114-12. Print 2012. mBio. 2012. PMID: 22807564 Free PMC article.
-
How Ralstonia solanacearum Exploits and Thrives in the Flowing Plant Xylem Environment.Trends Microbiol. 2018 Nov;26(11):929-942. doi: 10.1016/j.tim.2018.06.002. Epub 2018 Jun 22. Trends Microbiol. 2018. PMID: 29941188 Review.
-
Getting to the root of Ralstonia invasion.Semin Cell Dev Biol. 2023 Oct-Nov;148-149:3-12. doi: 10.1016/j.semcdb.2022.12.002. Epub 2022 Dec 14. Semin Cell Dev Biol. 2023. PMID: 36526528 Review.
Cited by
-
Combination of Bacillus and Low Fertigation Input Promoted the Growth and Productivity of Chinese Cabbage and Enriched Beneficial Rhizosphere Bacteria Lechevalieria.Biology (Basel). 2023 Aug 14;12(8):1130. doi: 10.3390/biology12081130. Biology (Basel). 2023. PMID: 37627014 Free PMC article.
-
Transcriptome Profiling Reveals the EanI/R Quorum Sensing Regulon in Pantoea Ananatis LMG 2665T.Genes (Basel). 2018 Mar 7;9(3):148. doi: 10.3390/genes9030148. Genes (Basel). 2018. PMID: 29518982 Free PMC article.
-
Transcriptomes of Ralstonia solanacearum during Root Colonization of Solanum commersonii.Front Plant Sci. 2017 Mar 20;8:370. doi: 10.3389/fpls.2017.00370. eCollection 2017. Front Plant Sci. 2017. PMID: 28373879 Free PMC article.
-
Plant-Pathogenic Ralstonia Phylotypes Evolved Divergent Respiratory Strategies and Behaviors To Thrive in Xylem.mBio. 2023 Feb 28;14(1):e0318822. doi: 10.1128/mbio.03188-22. Epub 2023 Feb 6. mBio. 2023. PMID: 36744950 Free PMC article.
-
Taxonomy and Phylogenetic Research on Ralstonia solanacearum Species Complex: A Complex Pathogen with Extraordinary Economic Consequences.Pathogens. 2020 Oct 25;9(11):886. doi: 10.3390/pathogens9110886. Pathogens. 2020. PMID: 33113847 Free PMC article. Review.
References
-
- Cornwell JC, Kemp WM, Kana TM. 1999. Denitrification in coastal ecosystems: methods, environmental controls, and ecosystem level controls, a review. Aquat Ecol 33:41–54. doi:10.1023/A:1009921414151. - DOI
-
- Kirchman DL. 2012. Processes in microbial ecology. Oxford University Press, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
