Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells
- PMID: 25784758
- PMCID: PMC4428043
- DOI: 10.15252/embr.201439789
Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells
Abstract
In mammals, the nuclear lamina interacts with hundreds of large genomic regions, termed lamina-associated domains (LADs) that are generally in a transcriptionally repressed state. Lamins form the major structural component of the lamina and have been reported to bind DNA and chromatin. Here, we systematically evaluate whether lamins are necessary for the LAD organization in murine embryonic stem cells. Surprisingly, removal of essentially all lamins does not have any detectable effect on the genome-wide interaction pattern of chromatin with emerin, a marker of the inner nuclear membrane. This suggests that other components of the lamina mediate these interactions.
Keywords: genome‐wide mapping; nuclear architecture; interphase chromosome organization; nuclear lamina.
© 2015 The Authors.
Figures
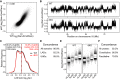
Scatterplot of log2 ratio of Dam-Emd over Dam for wt compared to dKO mES cells. Pearson correlation coefficient (r) is indicated.
Emd interaction profile along chromosome 18 in wt and dKO mES cells.
Distribution of LAD sizes in wt (black) and dKO (red) mES cells. Dashed lines mark the median LAD sizes. Percentage of total genomic DNA covered by LADs (% of gDNA), the median size of LADs, and the total number of LADs are listed for wt (black) and dKO (red) cells.
Concordance between wt and dKO cells for LADs and iLADs as defined in (C).
Distribution of Emd DamID probe values in wt and dKO mES cells, divided into facultative (f) and constitutive (c) LADs and iLADs. Horizontal lines of boxes depict percentiles 25, 50, and 75; vertical lines extend from the box edge to the highest or lowest value that is within 1.5× inter-quartile range of the edge.
Concordance between wt and dKO cells for facultative (fLAD, fiLAD) and constitutive (cLAD, ciLAD) regions, as identified in (E).

A, B Density scatterplot of the mean Emd DamID score per gene versus expression levels based on RNA-seq for wt (A) and dKO (B) mES cells.
C Density scatterplot of changes in Emd interaction versus changes in gene expression in dKO relative to wt cells.
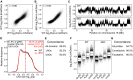
A–C Comparisons of LmA and Emd interaction profiles in wt and dKO mES cells. Samples were smoothed (median) with a running window of 11 probes. Pearson correlation coefficients (r) are indicated.
D–G Analysis of LADs as detected with Dam-LmA in wt and dKO cells, similar to Fig1C–F, respectively.

A Scatterplot of log2 ratio of Dam-Emd over Dam for wt against tKO mES cells. Pearson correlation coefficient (r) is indicated.
B Emd interaction profile along chromosome 18 in wt and tKO mES cells.
C–F Analysis of LADs as detected with Dam-Emd in wt and tKO cells, similar to Fig1C–F, respectively.
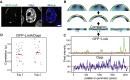
Representative confocal image showing the patchy NL localization of GFP-LmA in dKO mES cells and the corresponding DAPI staining of DNA. Scale bar, 5 μm.
Cartoon showing how the image analysis was performed. LmA patches are depicted in green, different DNA concentrations in shades of blue. The correlation between LmA and DAPI pixel intensity is determined in a 10-pixel shell, as shown in the bottom row.
Pixel intensity values of GFP-LmnA and DAPI along the cell perimeter for the nucleus shown (A).
Correlation analysis of DAPI and GFP-LmA signals along the nuclear perimeter. Graph shows the distribution of Spearman correlation coefficients for two independent experiments (Exp1, n = 31; Exp2, n = 23). These distributions are not significantly different from 0 (P > 0.2, Student's t-test). Red lines mark median correlation coefficients.
References
-
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. - PubMed
-
- Amendola M, van Steensel B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol. 2014;28:61–68. - PubMed
-
- Mendez-Lopez I, Worman HJ. Inner nuclear membrane proteins: impact on human disease. Chromosoma. 2012;121:153–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

