A novel host-proteome signature for distinguishing between acute bacterial and viral infections
- PMID: 25785720
- PMCID: PMC4364938
- DOI: 10.1371/journal.pone.0120012
A novel host-proteome signature for distinguishing between acute bacterial and viral infections
Abstract
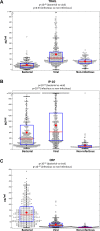
Bacterial and viral infections are often clinically indistinguishable, leading to inappropriate patient management and antibiotic misuse. Bacterial-induced host proteins such as procalcitonin, C-reactive protein (CRP), and Interleukin-6, are routinely used to support diagnosis of infection. However, their performance is negatively affected by inter-patient variability, including time from symptom onset, clinical syndrome, and pathogens. Our aim was to identify novel viral-induced host proteins that can complement bacterial-induced proteins to increase diagnostic accuracy. Initially, we conducted a bioinformatic screen to identify putative circulating host immune response proteins. The resulting 600 candidates were then quantitatively screened for diagnostic potential using blood samples from 1002 prospectively recruited patients with suspected acute infectious disease and controls with no apparent infection. For each patient, three independent physicians assigned a diagnosis based on comprehensive clinical and laboratory investigation including PCR for 21 pathogens yielding 319 bacterial, 334 viral, 112 control and 98 indeterminate diagnoses; 139 patients were excluded based on predetermined criteria. The best performing host-protein was TNF-related apoptosis-inducing ligand (TRAIL) (area under the curve [AUC] of 0.89; 95% confidence interval [CI], 0.86 to 0.91), which was consistently up-regulated in viral infected patients. We further developed a multi-protein signature using logistic-regression on half of the patients and validated it on the remaining half. The signature with the highest precision included both viral- and bacterial-induced proteins: TRAIL, Interferon gamma-induced protein-10, and CRP (AUC of 0.94; 95% CI, 0.92 to 0.96). The signature was superior to any of the individual proteins (P<0.001), as well as routinely used clinical parameters and their combinations (P<0.001). It remained robust across different physiological systems, times from symptom onset, and pathogens (AUCs 0.87-1.0). The accurate differential diagnosis provided by this novel combination of viral- and bacterial-induced proteins has the potential to improve management of patients with acute infections and reduce antibiotic misuse.
Conflict of interest statement
Figures

References
-
- Threat Report 2013 | Antimicrobial Resistance | CDC [Internet]. [cited 10 Nov 2013]. Available: http://www.cdc.gov/drugresistance/threat-report-2013/
-
- Craig JC, Williams GJ, Jones M, Codarini M, Macaskill P, Hayen A, et al. The accuracy of clinical symptoms and signs for the diagnosis of serious bacterial infection in young febrile children: prospective cohort study of 15 781 febrile illnesses. BMJ. 2010;340: c1594 10.1136/bmj.c1594 - DOI - PMC - PubMed
-
- Dedier J, Singer DE, Chang Y, Moore M, Atlas SJ. Processes of care, illness severity, and outcomes in the management of community-acquired pneumonia at academic hospitals. Arch Intern Med. 2001;161: 2099–2104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

