Novel R pipeline for analyzing Biolog Phenotypic MicroArray data
- PMID: 25786143
- PMCID: PMC4365023
- DOI: 10.1371/journal.pone.0118392
Novel R pipeline for analyzing Biolog Phenotypic MicroArray data
Abstract
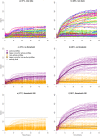
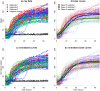
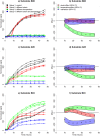
Data produced by Biolog Phenotype MicroArrays are longitudinal measurements of cells' respiration on distinct substrates. We introduce a three-step pipeline to analyze phenotypic microarray data with novel procedures for grouping, normalization and effect identification. Grouping and normalization are standard problems in the analysis of phenotype microarrays defined as categorizing bacterial responses into active and non-active, and removing systematic errors from the experimental data, respectively. We expand existing solutions by introducing an important assumption that active and non-active bacteria manifest completely different metabolism and thus should be treated separately. Effect identification, in turn, provides new insights into detecting differing respiration patterns between experimental conditions, e.g. between different combinations of strains and temperatures, as not only the main effects but also their interactions can be evaluated. In the effect identification, the multilevel data are effectively processed by a hierarchical model in the Bayesian framework. The pipeline is tested on a data set of 12 phenotypic plates with bacterium Yersinia enterocolitica. Our pipeline is implemented in R language on the top of opm R package and is freely available for research purposes.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

