A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea
- PMID: 25786576
- PMCID: PMC4365403
- DOI: 10.1038/srep09264
A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea
Abstract
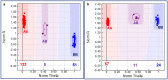
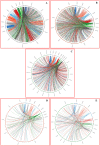
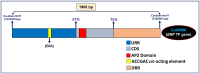
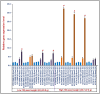
High experimental validation/genotyping success rate (94-96%) and intra-specific polymorphic potential (82-96%) of 1536 SNP and 472 SSR markers showing in silico polymorphism between desi ICC 4958 and kabuli ICC 12968 chickpea was obtained in a 190 mapping population (ICC 4958 × ICC 12968) and 92 diverse desi and kabuli genotypes. A high-density 2001 marker-based intra-specific genetic linkage map comprising of eight LGs constructed is comparatively much saturated (mean map-density: 0.94 cM) in contrast to existing intra-specific genetic maps in chickpea. Fifteen robust QTLs (PVE: 8.8-25.8% with LOD: 7.0-13.8) associated with pod and seed number/plant (PN and SN) and 100 seed weight (SW) were identified and mapped on 10 major genomic regions of eight LGs. One of 126.8 kb major genomic region harbouring a strong SW-associated robust QTL (Caq'SW1.1: 169.1-171.3 cM) has been delineated by integrating high-resolution QTL mapping with comprehensive marker-based comparative genome mapping and differential expression profiling. This identified one potential regulatory SNP (G/A) in the cis-acting element of candidate ERF (ethylene responsive factor) TF (transcription factor) gene governing seed weight in chickpea. The functionally relevant molecular tags identified have potential to be utilized for marker-assisted genetic improvement of chickpea.
Figures




References
-
- Winter P. et al. Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol. Gen. Genet. 262, 90–101 (1999). - PubMed
-
- Winter P. et al. A linkage map of chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum x C. reticulatum cross: localization of resistance genes for Fusarium wilt races 4 and 5. Theor. Appl. Genet. 101, 1155–1163 (2000).
-
- Abbo S. et al. Quantitative trait loci governing carotenoid concentration and weight in seed of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 111, 185–195 (2005). - PubMed
-
- Millan T. et al. A consensus genetic map of chickpea (Cicer arietinum L.) based on 10 mapping populations. Euphytica 175, 175–189 (2010).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

