KDM1 class flavin-dependent protein lysine demethylases
- PMID: 25787087
- PMCID: PMC4747437
- DOI: 10.1002/bip.22643
KDM1 class flavin-dependent protein lysine demethylases
Abstract
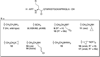
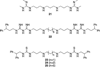
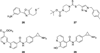
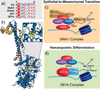
Flavin-dependent, lysine-specific protein demethylases (KDM1s) are a subfamily of amine oxidases that catalyze the selective posttranslational oxidative demethylation of methyllysine side chains within protein and peptide substrates. KDM1s participate in the widespread epigenetic regulation of both normal and disease state transcriptional programs. Their activities are central to various cellular functions, such as hematopoietic and neuronal differentiation, cancer proliferation and metastasis, and viral lytic replication and establishment of latency. Interestingly, KDM1s function as catalytic subunits within complexes with coregulatory molecules that modulate enzymatic activity of the demethylases and coordinate their access to specific substrates at distinct sites within the cell and chromatin. Although several classes of KDM1-selective small molecule inhibitors have been recently developed, these pan-active site inhibition strategies lack the ability to selectively discriminate between KDM1 activity in specific, and occasionally opposing, functional contexts within these complexes. Here we review the discovery of this class of demethylases, their structures, chemical mechanisms, and specificity. Additionally, we review inhibition of this class of enzymes as well as emerging interactions with coregulatory molecules that regulate demethylase activity in highly specific functional contexts of biological and potential therapeutic importance.
Keywords: Epigenetic; KDM1A; KDM1B; LSD1; LSD2.
© 2015 Wiley Periodicals, Inc.
Figures



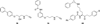





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

