Mapping Bias Overestimates Reference Allele Frequencies at the HLA Genes in the 1000 Genomes Project Phase I Data
- PMID: 25787242
- PMCID: PMC4426377
- DOI: 10.1534/g3.114.015784
Mapping Bias Overestimates Reference Allele Frequencies at the HLA Genes in the 1000 Genomes Project Phase I Data
Abstract
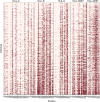
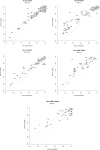
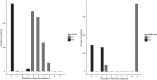
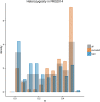
Next-generation sequencing (NGS) technologies have become the standard for data generation in studies of population genomics, as the 1000 Genomes Project (1000G). However, these techniques are known to be problematic when applied to highly polymorphic genomic regions, such as the human leukocyte antigen (HLA) genes. Because accurate genotype calls and allele frequency estimations are crucial to population genomics analyses, it is important to assess the reliability of NGS data. Here, we evaluate the reliability of genotype calls and allele frequency estimates of the single-nucleotide polymorphisms (SNPs) reported by 1000G (phase I) at five HLA genes (HLA-A, -B, -C, -DRB1, and -DQB1). We take advantage of the availability of HLA Sanger sequencing of 930 of the 1092 1000G samples and use this as a gold standard to benchmark the 1000G data. We document that 18.6% of SNP genotype calls in HLA genes are incorrect and that allele frequencies are estimated with an error greater than ±0.1 at approximately 25% of the SNPs in HLA genes. We found a bias toward overestimation of reference allele frequency for the 1000G data, indicating mapping bias is an important cause of error in frequency estimation in this dataset. We provide a list of sites that have poor allele frequency estimates and discuss the outcomes of including those sites in different kinds of analyses. Because the HLA region is the most polymorphic in the human genome, our results provide insights into the challenges of using of NGS data at other genomic regions of high diversity.
Keywords: 1000 Genomes; HLA; NGS; mapping bias.
Copyright © 2015 Brandt et al.
Figures



References
-
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., et al. , 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329: 506–512. - PubMed
-
- Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., et al. , 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39. - PubMed
-
- Chapman S. J., Hill A. V. S., 2012. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 13: 175–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
