Single cell visualization of transcription kinetics variance of highly mobile identical genes using 3D nanoimaging
- PMID: 25788248
- PMCID: PMC4365385
- DOI: 10.1038/srep09258
Single cell visualization of transcription kinetics variance of highly mobile identical genes using 3D nanoimaging
Abstract
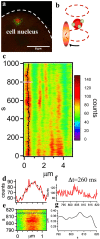
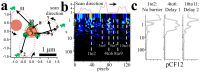
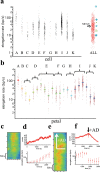
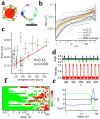
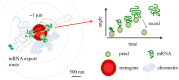
Multi-cell biochemical assays and single cell fluorescence measurements revealed that the elongation rate of Polymerase II (PolII) in eukaryotes varies largely across different cell types and genes. However, there is not yet a consensus whether intrinsic factors such as the position, local mobility or the engagement by an active molecular mechanism of a genetic locus could be the determinants of the observed heterogeneity. Here by employing high-speed 3D fluorescence nanoimaging techniques we resolve and track at the single cell level multiple, distinct regions of mRNA synthesis within the model system of a large transgene array. We demonstrate that these regions are active transcription sites that release mRNA molecules in the nucleoplasm. Using fluctuation spectroscopy and the phasor analysis approach we were able to extract the local PolII elongation rate at each site as a function of time. We measured a four-fold variation in the average elongation between identical copies of the same gene measured simultaneously within the same cell, demonstrating a correlation between local transcription kinetics and the movement of the transcription site. Together these observations demonstrate that local factors, such as chromatin local mobility and the microenvironment of the transcription site, are an important source of transcription kinetics variability.
Figures
References
-
- Ardehali M. B. & Lis J. T. Tracking rates of transcription and splicing in vivo. Nature structural & molecular biology 16, 1123–1124 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

