Constitutive heterochromatin formation and transcription in mammals
- PMID: 25788984
- PMCID: PMC4363358
- DOI: 10.1186/1756-8935-8-3
Constitutive heterochromatin formation and transcription in mammals
Abstract
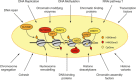
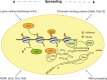
Constitutive heterochromatin, mainly formed at the gene-poor regions of pericentromeres, is believed to ensure a condensed and transcriptionally inert chromatin conformation. Pericentromeres consist of repetitive tandem satellite repeats and are crucial chromosomal elements that are responsible for accurate chromosome segregation in mitosis. The repeat sequences are not conserved and can greatly vary between different organisms, suggesting that pericentromeric functions might be controlled epigenetically. In this review, we will discuss how constitutive heterochromatin is formed and maintained at pericentromeres in order to ensure their integrity. We will describe the biogenesis and the function of main epigenetic pathways that are involved and how they are interconnected. Interestingly, recent findings suggest that alternative pathways could substitute for well-established pathways when disrupted, suggesting that constitutive heterochromatin harbors much more plasticity than previously assumed. In addition, despite of the heterochromatic nature of pericentromeres, there is increasing evidence for active and regulated transcription at these loci, in a multitude of organisms and under various biological contexts. Thus, in the second part of this review, we will address this relatively new aspect and discuss putative functions of pericentromeric expression.
Keywords: epigenetic factors; heterochromatin; histone modifying enzymes; pericentromere; transcription.
Figures

References
-
- Eymery A, Callanan M, Vourc’h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol. 2009;53(2–3):259–68. - PubMed
-
- Schueler MG, Sullivan BA. Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet. 2006;7:301–13. - PubMed
-
- Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol. 2010;26:471–501. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

