Stranglehold on the spindle assembly checkpoint: the human papillomavirus E2 protein provokes BUBR1-dependent aneuploidy
- PMID: 25789401
- PMCID: PMC4614697
- DOI: 10.1080/15384101.2015.1021519
Stranglehold on the spindle assembly checkpoint: the human papillomavirus E2 protein provokes BUBR1-dependent aneuploidy
Abstract
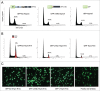
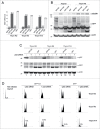
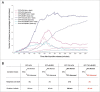
The Human Papillomavirus (HPV) E2 protein, which inhibits the E6 and E7 viral oncogenes, is believed to have anti-oncogenic properties. Here, we challenge this view and show that HPV-18 E2 over-activates the Spindle Assembly Checkpoint (SAC) and induces DNA breaks in mitosis followed by aneuploidy. This phenotype is associated with interaction of E2 with the Mitotic Checkpoint Complex (MCC) proteins Cdc20, MAD2 and BUBR1. While BUBR1 silencing rescues the mitotic phenotype induced by E2, p53 silencing or presence of E6/E7 (inactivating p53 and increasing BUBR1 levels respectively) both amplify it. This work pinpoints E2 as a key protein in the initiation of HPV-induced cervical cancer and identifies the SAC as a target for oncogenic pathogens. Moreover, our results suggest a role of p53 in regulating the mitotic process itself and highlight SAC over-activation in a p53-negative context as a highly pathogenic event.
Keywords: APC/C, Anaphase Promoting Complex/Cyclosome; Ad, Adenovirus; BUBR1; E2; E2 TAD, E2 Transactivation Domain; E2 ΔTAD, E2 deleted of the Transactivation Domain; GFP, Green Fluorescent Protein; HPV, Human Papillomavirus; MCC, Mitotic Checkpoint Complex; MS, Mass Spectrometry; Noco, Nocodazole; SAC, Spindle Assembly Checkpoint; Thym, Thymidine; aneuploidy; m.o.i., Multiplicity of Infection; mitosis; p53; papillomavirus; spindle assembly checkpoint.
Figures



Similar articles
-
Role of ubiquitylation of components of mitotic checkpoint complex in their dissociation from anaphase-promoting complex/cyclosome.Proc Natl Acad Sci U S A. 2018 Feb 20;115(8):1777-1782. doi: 10.1073/pnas.1720312115. Epub 2018 Feb 5. Proc Natl Acad Sci U S A. 2018. PMID: 29432156 Free PMC article.
-
Compromised spindle assembly checkpoint due to altered expression of Ubch10 and Cdc20 in human papillomavirus type 16 E6- and E7-expressing keratinocytes.J Virol. 2010 Nov;84(21):10956-64. doi: 10.1128/JVI.00259-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739533 Free PMC article.
-
Monopolar spindle 1 (MPS1) kinase promotes production of closed MAD2 (C-MAD2) conformer and assembly of the mitotic checkpoint complex.J Biol Chem. 2013 Dec 6;288(49):35149-58. doi: 10.1074/jbc.M113.522375. Epub 2013 Oct 22. J Biol Chem. 2013. PMID: 24151075 Free PMC article.
-
The regulation of cell proliferation by the papillomavirus early proteins.Cell Mol Life Sci. 2009 May;66(10):1700-17. doi: 10.1007/s00018-009-8631-7. Cell Mol Life Sci. 2009. PMID: 19183849 Free PMC article. Review.
-
Aneuploidy and tumorigenesis.Semin Cell Dev Biol. 2011 Aug;22(6):595-601. doi: 10.1016/j.semcdb.2011.03.002. Epub 2011 Mar 15. Semin Cell Dev Biol. 2011. PMID: 21392584 Free PMC article. Review.
Cited by
-
Environmental Stress-Induced Alterations in Embryo Developmental Morphokinetics.J Xenobiot. 2024 Oct 21;14(4):1613-1637. doi: 10.3390/jox14040087. J Xenobiot. 2024. PMID: 39449428 Free PMC article. Review.
-
Hitchhiking of Viral Genomes on Cellular Chromosomes.Annu Rev Virol. 2019 Sep 29;6(1):275-296. doi: 10.1146/annurev-virology-092818-015716. Epub 2019 Jul 5. Annu Rev Virol. 2019. PMID: 31283444 Free PMC article. Review.
-
HPV16-E2 induces prophase arrest and activates the cellular DNA damage response in vitro and in precursor lesions of cervical carcinoma.Oncotarget. 2015 Oct 27;6(33):34979-91. doi: 10.18632/oncotarget.5512. Oncotarget. 2015. PMID: 26474276 Free PMC article.
-
Bioinformatics analysis shows that TOP2A functions as a key candidate gene in the progression of cervical cancer.Biomed Rep. 2020 Oct;13(4):21. doi: 10.3892/br.2020.1328. Epub 2020 Jul 9. Biomed Rep. 2020. PMID: 32765860 Free PMC article.
-
HPV-18 E2 protein downregulates antisense noncoding mitochondrial RNA-2, delaying replicative senescence of human keratinocytes.Aging (Albany NY). 2018 Dec 30;11(1):33-47. doi: 10.18632/aging.101711. Aging (Albany NY). 2018. PMID: 30595560 Free PMC article.
References
-
- zur Hausen H. Condylomata acuminata and human genital cancer. Cancer Res 1976; 36: 794; PMID:175942 - PubMed
-
- Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet 2007; 370: 890-907; PMID:17826171; http://dx.doi.org/10.1016/S0140-6736(07)61416-0 - DOI - PubMed
-
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990; 63: 1129-36; PMID:2175676; http://dx.doi.org/10.1016/0092-8674(90)90409-8 - DOI - PubMed
-
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990; 248: 76-9; PMID:2157286; http://dx.doi.org/10.1126/science.2157286 - DOI - PubMed
-
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV16 E6 and E6-AP complex functions as ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993; 75: 495-505; PMID:8221889; http://dx.doi.org/10.1016/0092-8674(93)90384-3 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
