Gene-based mapping and pathway analysis of metabolic traits in dairy cows
- PMID: 25789767
- PMCID: PMC4366076
- DOI: 10.1371/journal.pone.0122325
Gene-based mapping and pathway analysis of metabolic traits in dairy cows
Abstract
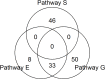
The metabolic adaptation of dairy cows during the transition period has been studied intensively in the last decades. However, until now, only few studies have paid attention to the genetic aspects of this process. Here, we present the results of a gene-based mapping and pathway analysis with the measurements of three key metabolites, (1) non-esterified fatty acids (NEFA), (2) beta-hydroxybutyrate (BHBA) and (3) glucose, characterizing the metabolic adaptability of dairy cows before and after calving. In contrast to the conventional single-marker approach, we identify 99 significant and biologically sensible genes associated with at least one of the considered phenotypes and thus giving evidence for a genetic basis of the metabolic adaptability. Moreover, our results strongly suggest three pathways involved in the metabolism of steroids and lipids are potential candidates for the adaptive regulation of dairy cows in their early lactation. From our perspective, a closer investigation of our findings will lead to a step forward in understanding the variability in the metabolic adaptability of dairy cows in their early lactation.
Conflict of interest statement
Figures

Similar articles
-
Comparison of hepatic adaptation in extreme metabolic phenotypes observed in early lactation dairy cows on-farm.J Anim Physiol Anim Nutr (Berl). 2014 Aug;98(4):693-703. doi: 10.1111/jpn.12125. Epub 2013 Sep 3. J Anim Physiol Anim Nutr (Berl). 2014. PMID: 24033645
-
Genetic relationship of body energy and blood metabolites with reproduction in holstein cows.J Dairy Sci. 2008 Nov;91(11):4323-32. doi: 10.3168/jds.2008-1018. J Dairy Sci. 2008. PMID: 18946138
-
The association between metabolic parameters and oocyte quality early and late postpartum in Holstein dairy cows.J Dairy Sci. 2012 Mar;95(3):1257-66. doi: 10.3168/jds.2011-4649. J Dairy Sci. 2012. PMID: 22365209
-
Elevated non-esterified fatty acids and β-hydroxybutyrate and their association with transition dairy cow performance.Vet J. 2013 Dec;198(3):560-70. doi: 10.1016/j.tvjl.2013.08.011. Epub 2013 Aug 16. Vet J. 2013. PMID: 24054909 Review.
-
Effects of dietary energy source on energy balance, metabolites and reproduction variables in dairy cows in early lactation.Theriogenology. 2007 Sep 1;68 Suppl 1:S274-80. doi: 10.1016/j.theriogenology.2007.04.043. Epub 2007 May 29. Theriogenology. 2007. PMID: 17537496 Review.
Cited by
-
The 'heritability' of domestication and its functional partitioning in the pig.Heredity (Edinb). 2017 Feb;118(2):160-168. doi: 10.1038/hdy.2016.78. Epub 2016 Sep 21. Heredity (Edinb). 2017. PMID: 27649617 Free PMC article.
-
Analysis of hepatic transcript profile and plasma lipid profile in early lactating dairy cows fed grape seed and grape marc meal extract.BMC Genomics. 2017 Mar 23;18(1):253. doi: 10.1186/s12864-017-3638-1. BMC Genomics. 2017. PMID: 28335726 Free PMC article.
-
A Pathway-Centered Analysis of Pig Domestication and Breeding in Eurasia.G3 (Bethesda). 2017 Jul 5;7(7):2171-2184. doi: 10.1534/g3.117.042671. G3 (Bethesda). 2017. PMID: 28500056 Free PMC article.
-
Longitudinal Phenotypes Improve Genotype Association for Hyperketonemia in Dairy Cattle.Animals (Basel). 2019 Dec 1;9(12):1059. doi: 10.3390/ani9121059. Animals (Basel). 2019. PMID: 31805754 Free PMC article.
-
Exploring the Genetic Basis of Ketosis: Preliminary GWAS Findings on Beta-Hydroxybutyrate Levels in Holstein Cattle.Int J Genomics. 2025 Jul 1;2025:5520648. doi: 10.1155/ijog/5520648. eCollection 2025. Int J Genomics. 2025. PMID: 40662169 Free PMC article.
References
-
- Wellnitz O, Bruckmaier R, Blum J. Milk ejection and milk removal of single quarters in high yielding dairy cows. Milk Sci Int. 1999;54.
-
- Bobe G, Young JW, Beitz DC. Invited review: pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J Dairy Sci. 2004;87: 3105–3124. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

