QSLiMFinder: improved short linear motif prediction using specific query protein data
- PMID: 25792551
- PMCID: PMC4495300
- DOI: 10.1093/bioinformatics/btv155
QSLiMFinder: improved short linear motif prediction using specific query protein data
Abstract
Motivation: The sensitivity of de novo short linear motif (SLiM) prediction is limited by the number of patterns (the motif space) being assessed for enrichment. QSLiMFinder uses specific query protein information to restrict the motif space and thereby increase the sensitivity and specificity of predictions.
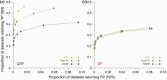
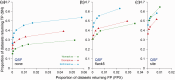
Results: QSLiMFinder was extensively benchmarked using known SLiM-containing proteins and simulated protein interaction datasets of real human proteins. Exploiting prior knowledge of a query protein likely to be involved in a SLiM-mediated interaction increased the proportion of true positives correctly returned and reduced the proportion of datasets returning a false positive prediction. The biggest improvement was seen if a short region of the query protein flanking the interaction site was known.
Availability and implementation: All the tools and data used in this study, including QSLiMFinder and the SLiMBench benchmarking software, are freely available under a GNU license as part of SLiMSuite, at: http://bioware.soton.ac.uk.
© The Author 2015. Published by Oxford University Press.
Figures





References
-
- Babu M.M., et al. (2011) Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol., 21, 432–440. - PubMed
-
- Bruning J.B., Shamoo Y. (2004) Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure, 12, 2209–2219. - PubMed
-
- Davey N.E., et al. . (2009) Masking residues using context-specific evolutionary conservation significantly improves short linear motif discovery. Bioinformatics, 25, 443–450. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

