A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway
- PMID: 25792599
- PMCID: PMC4378196
- DOI: 10.1101/gad.258731.115
A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway
Abstract
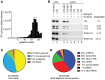
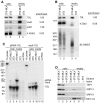
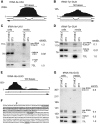
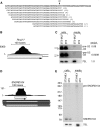
Although all retroviruses recruit host cell RNAs into virions, both the spectrum of RNAs encapsidated and the mechanisms by which they are recruited remain largely unknown. Here, we used high-throughput sequencing to obtain a comprehensive description of the RNAs packaged by a model retrovirus, murine leukemia virus. The major encapsidated host RNAs are noncoding RNAs (ncRNAs) and members of the VL30 class of endogenous retroviruses. Remarkably, although Moloney leukemia virus (MLV) assembles in the cytoplasm, precursors to specific tRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) are all enriched in virions. Consistent with their cytoplasmic recruitment, packaging of both pre-tRNAs and U6 snRNA requires the nuclear export receptor Exportin-5. Adenylated and uridylated forms of these RNAs accumulate in cells and virions when the cytoplasmic exoribonuclease DIS3L2 and subunits of the RNA exosome are depleted. Together, our data reveal that MLV recruits RNAs from a novel host cell surveillance pathway in which unprocessed and unneeded nuclear ncRNAs are exported to the cytoplasm for degradation.
Keywords: RNA surveillance; exoribonucleases; noncoding RNAs; retrovirus.
© 2015 Eckwahl et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr Top Microbiol Immunol 214: 177–218. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
