Evidence for widespread subfunctionalization of splice forms in vertebrate genomes
- PMID: 25792610
- PMCID: PMC4417111
- DOI: 10.1101/gr.184473.114
Evidence for widespread subfunctionalization of splice forms in vertebrate genomes
Abstract
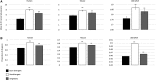
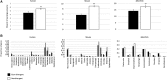
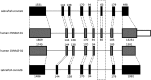
Gene duplication and alternative splicing are important sources of proteomic diversity. Despite research indicating that gene duplication and alternative splicing are negatively correlated, the evolutionary relationship between the two remains unclear. One manner in which alternative splicing and gene duplication may be related is through the process of subfunctionalization, in which an alternatively spliced gene upon duplication divides distinct splice isoforms among the newly generated daughter genes, in this way reducing the number of alternatively spliced transcripts duplicate genes produce. Previously, it has been shown that splice form subfunctionalization will result in duplicate pairs with divergent exon structure when distinct isoforms become fixed in each paralog. However, the effects of exon structure divergence between paralogs have never before been studied on a genome-wide scale. Here, using genomic data from human, mouse, and zebrafish, we demonstrate that gene duplication followed by exon structure divergence between paralogs results in a significant reduction in levels of alternative splicing. In addition, by comparing the exon structure of zebrafish duplicates to the co-orthologous human gene, we have demonstrated that a considerable fraction of exon divergent duplicates maintain the structural signature of splice form subfunctionalization. Furthermore, we find that paralogs with divergent exon structure demonstrate reduced breadth of expression in a variety of tissues when compared to paralogs with identical exon structures and singletons. Taken together, our results are consistent with subfunctionalization partitioning alternatively spliced isoforms among duplicate genes and as such highlight the relationship between gene duplication and alternative splicing.
© 2015 Lambert et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


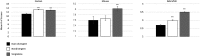
Similar articles
-
The evolutionary fate of alternatively spliced homologous exons after gene duplication.Genome Biol Evol. 2015 Apr 29;7(6):1392-403. doi: 10.1093/gbe/evv076. Genome Biol Evol. 2015. PMID: 25931610 Free PMC article.
-
Gene duplication followed by exon structure divergence substitutes for alternative splicing in zebrafish.Gene. 2014 Aug 10;546(2):271-6. doi: 10.1016/j.gene.2014.05.068. Epub 2014 Jun 3. Gene. 2014. PMID: 24942242
-
Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis.Mol Biol Evol. 2010 Jul;27(7):1686-97. doi: 10.1093/molbev/msq054. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185454
-
Splicing for alternative structures of Cav1.2 Ca2+ channels in cardiac and smooth muscles.Cardiovasc Res. 2005 Nov 1;68(2):197-203. doi: 10.1016/j.cardiores.2005.06.024. Epub 2005 Jul 27. Cardiovasc Res. 2005. PMID: 16051206 Review.
-
Alternative splicing and the evolution of phenotypic novelty.Philos Trans R Soc Lond B Biol Sci. 2017 Feb 5;372(1713):20150474. doi: 10.1098/rstb.2015.0474. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 27994117 Free PMC article. Review.
Cited by
-
Similarities in biological processes can be used to bridge ecology and molecular biology.Evol Appl. 2020 Apr 13;13(6):1335-1350. doi: 10.1111/eva.12961. eCollection 2020 Jul. Evol Appl. 2020. PMID: 32684962 Free PMC article.
-
A targeted gene expression platform allows for rapid analysis of chemical-induced antioxidant mRNA expression in zebrafish larvae.PLoS One. 2017 Feb 17;12(2):e0171025. doi: 10.1371/journal.pone.0171025. eCollection 2017. PLoS One. 2017. PMID: 28212397 Free PMC article.
-
Zebrafish Models of Rare Hereditary Pediatric Diseases.Diseases. 2018 May 22;6(2):43. doi: 10.3390/diseases6020043. Diseases. 2018. PMID: 29789451 Free PMC article. Review.
-
Evolutionary and Functional Analysis of Membrane-Bound NAC Transcription Factor Genes in Soybean.Plant Physiol. 2016 Nov;172(3):1804-1820. doi: 10.1104/pp.16.01132. Epub 2016 Sep 26. Plant Physiol. 2016. PMID: 27670816 Free PMC article.
-
The evolutionary fate of alternatively spliced homologous exons after gene duplication.Genome Biol Evol. 2015 Apr 29;7(6):1392-403. doi: 10.1093/gbe/evv076. Genome Biol Evol. 2015. PMID: 25931610 Free PMC article.
References
-
- Arjona FJ, Chen YX, Flik G, Bindels RJ, Hoenderop JG. 2013. Tissue-specific expression and in vivo regulation of zebrafish orthologues of mammalian genes related to symptomatic hypomagnesemia. Pflugers Arch 465: 1409–1421. - PubMed
-
- Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
