Neurospora importin α is required for normal heterochromatic formation and DNA methylation
- PMID: 25793375
- PMCID: PMC4368784
- DOI: 10.1371/journal.pgen.1005083
Neurospora importin α is required for normal heterochromatic formation and DNA methylation
Abstract
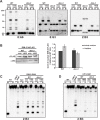
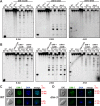
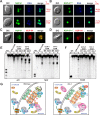
Heterochromatin and associated gene silencing processes play roles in development, genome defense, and chromosome function. In many species, constitutive heterochromatin is decorated with histone H3 tri-methylated at lysine 9 (H3K9me3) and cytosine methylation. In Neurospora crassa, a five-protein complex, DCDC, catalyzes H3K9 methylation, which then directs DNA methylation. Here, we identify and characterize a gene important for DCDC function, dim-3 (defective in methylation-3), which encodes the nuclear import chaperone NUP-6 (Importin α). The critical mutation in dim-3 results in a substitution in an ARM repeat of NUP-6 and causes a substantial loss of H3K9me3 and DNA methylation. Surprisingly, nuclear transport of all known proteins involved in histone and DNA methylation, as well as a canonical transport substrate, appear normal in dim-3 strains. Interactions between DCDC members also appear normal, but the nup-6(dim-3) allele causes the DCDC members DIM-5 and DIM-7 to mislocalize from heterochromatin and NUP-6dim-3 itself is mislocalized from the nuclear envelope, at least in conidia. GCN-5, a member of the SAGA histone acetyltransferase complex, also shows altered localization in dim-3, raising the possibility that NUP-6 is necessary to localize multiple chromatin complexes following nucleocytoplasmic transport.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC.PLoS Genet. 2010 Nov 4;6(11):e1001196. doi: 10.1371/journal.pgen.1001196. PLoS Genet. 2010. PMID: 21079689 Free PMC article.
-
Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9598-E9607. doi: 10.1073/pnas.1715049114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078403 Free PMC article.
-
Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3.Genome Res. 2016 Aug;26(8):1069-80. doi: 10.1101/gr.203182.115. Epub 2016 Jun 3. Genome Res. 2016. PMID: 27260477 Free PMC article.
-
DNA methylation and the formation of heterochromatin in Neurospora crassa.Heredity (Edinb). 2010 Jul;105(1):38-44. doi: 10.1038/hdy.2010.44. Epub 2010 Apr 21. Heredity (Edinb). 2010. PMID: 20407471 Review.
-
[On the research of histone methylation].Yi Chuan. 2004 Mar;26(2):244-8. Yi Chuan. 2004. PMID: 15639996 Review. Chinese.
Cited by
-
Opposing functions of Fng1 and the Rpd3 HDAC complex in H4 acetylation in Fusarium graminearum.PLoS Genet. 2020 Nov 2;16(11):e1009185. doi: 10.1371/journal.pgen.1009185. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137093 Free PMC article.
-
Direct observation of importin α family member KPNA1 in axonal transport with or without a schizophrenia-related mutation.J Biol Chem. 2025 Apr;301(4):108343. doi: 10.1016/j.jbc.2025.108343. Epub 2025 Feb 24. J Biol Chem. 2025. PMID: 40010609 Free PMC article.
-
Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):15048-15053. doi: 10.1073/pnas.1615546113. Epub 2016 Nov 16. Proc Natl Acad Sci U S A. 2016. PMID: 27856763 Free PMC article.
-
A Matter of Scale and Dimensions: Chromatin of Chromosome Landmarks in the Fungi.Microbiol Spectr. 2017 Jul;5(4):10.1128/microbiolspec.funk-0054-2017. doi: 10.1128/microbiolspec.FUNK-0054-2017. Microbiol Spectr. 2017. PMID: 28752814 Free PMC article. Review.
-
SWO1 modulates cell wall integrity under salt stress by interacting with importin ɑ in Arabidopsis.Stress Biol. 2021 Sep 29;1(1):9. doi: 10.1007/s44154-021-00010-5. Stress Biol. 2021. PMID: 37676567 Free PMC article.
References
-
- Henikoff S (2000) Heterochromatin function in complex genomes. Biochim Biophys Acta 1470: O1–8. - PubMed
-
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

