Mid-embryo patterning and precision in Drosophila segmentation: Krüppel dual regulation of hunchback
- PMID: 25793381
- PMCID: PMC4368514
- DOI: 10.1371/journal.pone.0118450
Mid-embryo patterning and precision in Drosophila segmentation: Krüppel dual regulation of hunchback
Abstract
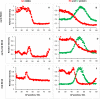
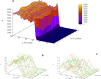
In early development, genes are expressed in spatial patterns which later define cellular identities and tissue locations. The mechanisms of such pattern formation have been studied extensively in early Drosophila (fruit fly) embryos. The gap gene hunchback (hb) is one of the earliest genes to be expressed in anterior-posterior (AP) body segmentation. As a transcriptional regulator for a number of downstream genes, the spatial precision of hb expression can have significant effects in the development of the body plan. To investigate the factors contributing to hb precision, we used fine spatial and temporal resolution data to develop a quantitative model for the regulation of hb expression in the mid-embryo. In particular, modelling hb pattern refinement in mid nuclear cleavage cycle 14 (NC14) reveals some of the regulatory contributions of simultaneously-expressed gap genes. Matching the model to recent data from wild-type (WT) embryos and mutants of the gap gene Krüppel (Kr) indicates that a mid-embryo Hb concentration peak important in thoracic development (at parasegment 4, PS4) is regulated in a dual manner by Kr, with low Kr concentration activating hb and high Kr concentration repressing hb. The processes of gene expression (transcription, translation, transport) are intrinsically random. We used stochastic simulations to characterize the noise generated in hb expression. We find that Kr regulation can limit the positional variability of the Hb mid-embryo border. This has been recently corroborated in experimental comparisons of WT and Kr- mutant embryos. Further, Kr regulation can decrease uncertainty in mid-embryo hb expression (i.e. contribute to a smooth Hb boundary) and decrease between-copy transcriptional variability within nuclei. Since many tissue boundaries are first established by interactions between neighbouring gene expression domains, these properties of Hb-Kr dynamics to diminish the effects of intrinsic expression noise may represent a general mechanism contributing to robustness in early development.
Conflict of interest statement
Figures




References
-
- Wolpert L. Positional information and spatial pattern of cellular differentiation. J Theor Biol. 1969;25: 1–47. - PubMed
-
- Ephrussi A, St Johnston D. Seeing is believing: The bicoid morphogen gradient matures. Cell 2004;116: 143–152. - PubMed
-
- Hülskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Krüppel and knirps in the early Drosophila embryo. Nature 1990;346: 577–580. - PubMed
-
- Bonneton F, Shaw PJ, Fazakerley C, Shi M, Dover GA. Comparison of bicoid-dependent regulation of hunchback between Musca domestica and Drosophila melanogaster . Mech Dev. 1997;66: 143–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

