Transcriptional regulation of tenascin genes
- PMID: 25793574
- PMCID: PMC4422812
- DOI: 10.1080/19336918.2015.1008333
Transcriptional regulation of tenascin genes
Abstract
Extracellular matrix proteins of the tenascin family resemble each other in their domain structure, and also share functions in modulating cell adhesion and cellular responses to growth factors. Despite these common features, the 4 vertebrate tenascins exhibit vastly different expression patterns. Tenascin-R is specific to the central nervous system. Tenascin-C is an "oncofetal" protein controlled by many stimuli (growth factors, cytokines, mechanical stress), but with restricted occurrence in space and time. In contrast, tenascin-X is a constituitive component of connective tissues, and its level is barely affected by external factors. Finally, the expression of tenascin-W is similar to that of tenascin-C but even more limited. In accordance with their highly regulated expression, the promoters of the tenascin-C and -W genes contain TATA boxes, whereas those of the other 2 tenascins do not. This article summarizes what is currently known about the complex transcriptional regulation of the 4 tenascin genes in development and disease.
Keywords: AKT, v-akt murine thymoma viral oncogene homolog; ALK, anaplastic lymphoma kinase; AP-1, activator protein-1; ATF, activating transcription factor; BMP, bone morphogenetic protein; CBP, CREB binding protein; CREB, cAMP response element-binding protein; CREB-RP, CREB-related protein; CYP21A2, cytochrome P450 family 21 subfamily A polypeptide 2; ChIP, chromatin immunoprecipitation; EBS, Ets binding site; ECM, extracellular matrix; EGF, epidermal growth factor; ERK1/2, extracellular signal-regulated kinase 1/2; ETS, E26 transformation-specific; EWS-ETS, Ewing sarcoma-Ets fusion protein; Evx1, even skipped homeobox 1; FGF, fibroblast growth factor; HBS, homeodomain binding sequence; IL, interleukin; ILK, integrin-linked kinase; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; MHCIII, major histocompatibility complex class III; MKL1, megakaryoblastic leukemia-1; NFκB, nuclear factor kappa B; NGF, nerve growth factor; NFAT, nuclear factor of activated T-cells; OTX2, orthodenticle homolog 2; PDGF, platelet-derived growth factor; PI3K, phosphatidylinositol 3-kinase; POU3F2, POU domain class 3 transcription factor 2; PRRX1, paired-related homeobox 1; RBPJk, recombining binding protein suppressor of hairless; ROCK, Rho-associated, coiled-coil-containing protein kinase; RhoA, ras homolog gene family member A; SAP, SAF-A/B, Acinus, and PIAS; SCX, scleraxix; SEAP, secreted alkaline phosphatase; SMAD, small body size - mothers against decapentaplegic; SOX4, sex determining region Y-box 4; SRE, serum response element; SRF, serum response factor; STAT, signal transducer and activator of transcription; TGF-β, transforming growth factor-β; TNC, tenascin-C; TNF-α, tumor necrosis factor-α; TNR, tenascin-R; TNW, tenascin-W; TNX, tenascin-X; TSS, transcription start site; UTR, untranslated region; WNT, wingless-related integration site; cancer; cytokine; development; extracellular matrix; gene promoter; gene regulation; glucocorticoid; growth factor; homeobox gene; matricellular; mechanical stress; miR, micro RNA; p38 MAPK, p38 mitogen activated protein kinase; tenascin; transcription factor.
Figures
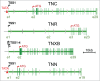
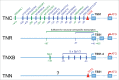
References
-
- Chiquet-Ehrismann R, Chiquet M. Tenascins: regulation and putative functions during pathological stress. J Pathol 2003; 200:488-99; PMID:12845616; http://dx.doi.org/ 10.1002/path.1415 - DOI - PubMed
-
- Tucker RP, Chiquet-Ehrismann R. Evidence for the evolution of tenascin and fibronectin early in the chordate lineage. Int J Biochem Cell Biol 2009; 41:424-34; PMID:18761101; http://dx.doi.org/ 10.1016/j.biocel.2008.08.003 - DOI - PubMed
-
- Murphy-Ullrich JE, Sage EH. Revisiting the matricellular concept. Matrix Biol 2014; 37:1-14; PMID:25064829; http://dx.doi.org/ 10.1016/j.matbio.2014.07.005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
