Molecular dynamics simulations of acylpeptide hydrolase bound to chlorpyrifosmethyl oxon and dichlorvos
- PMID: 25794283
- PMCID: PMC4394528
- DOI: 10.3390/ijms16036217
Molecular dynamics simulations of acylpeptide hydrolase bound to chlorpyrifosmethyl oxon and dichlorvos
Abstract
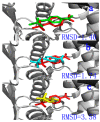
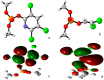
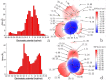
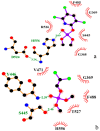
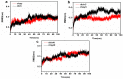
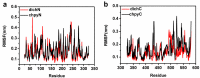
Acylpeptide hydrolases (APHs) catalyze the removal of N-acylated amino acids from blocked peptides. Like other prolyloligopeptidase (POP) family members, APHs are believed to be important targets for drug design. To date, the binding pose of organophosphorus (OP) compounds of APH, as well as the different OP compounds binding and inducing conformational changes in two domains, namely, α/β hydrolase and β-propeller, remain poorly understood. We report a computational study of APH bound to chlorpyrifosmethyl oxon and dichlorvos. In our docking study, Val471 and Gly368 are important residues for chlorpyrifosmethyl oxon and dichlorvos binding. Molecular dynamics simulations were also performed to explore the conformational changes between the chlorpyrifosmethyl oxon and dichlorvos bound to APH, which indicated that the structural feature of chlorpyrifosmethyl oxon binding in APH permitted partial opening of the β-propeller fold and allowed the chlorpyrifosmethyl oxon to easily enter the catalytic site. These results may facilitate the design of APH-targeting drugs with improved efficacy.
Figures
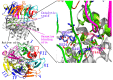

Similar articles
-
Exploration of the chlorpyrifos escape pathway from acylpeptide hydrolases using steered molecular dynamics simulations.J Biomol Struct Dyn. 2016;34(4):749-61. doi: 10.1080/07391102.2015.1052097. Epub 2015 Jul 8. J Biomol Struct Dyn. 2016. PMID: 26155973
-
Identification of acylpeptide hydrolase as a sensitive site for reaction with organophosphorus compounds and a potential target for cognitive enhancing drugs.Mol Pharmacol. 2000 Sep;58(3):577-83. doi: 10.1124/mol.58.3.577. Mol Pharmacol. 2000. PMID: 10953051
-
Blood acylpeptide hydrolase activity is a sensitive marker for exposure to some organophosphate toxicants.Toxicol Sci. 2005 Aug;86(2):291-9. doi: 10.1093/toxsci/kfi195. Epub 2005 May 11. Toxicol Sci. 2005. PMID: 15888665
-
Catabolism of intracellular N-terminal acetylated proteins: involvement of acylpeptide hydrolase and acylase.Biochimie. 2005 Aug;87(8):673-85. doi: 10.1016/j.biochi.2005.04.002. Biochimie. 2005. PMID: 15927344 Review.
-
Noncholinesterase effects induced by organophosphate pesticides and their relationship to cognitive processes: implication for the action of acylpeptide hydrolase.J Toxicol Environ Health B Crit Rev. 2007 Dec;10(8):623-30. doi: 10.1080/10937400701436445. J Toxicol Environ Health B Crit Rev. 2007. PMID: 18049927 Review.
Cited by
-
Exploration of binding and inhibition mechanism of a small molecule inhibitor of influenza virus H1N1 hemagglutinin by molecular dynamics simulation.Sci Rep. 2017 Jun 19;7(1):3786. doi: 10.1038/s41598-017-03719-4. Sci Rep. 2017. PMID: 28630402 Free PMC article.
-
Synthesis of Novel Benzothiazole-Profen Hybrid Amides as Potential NSAID Candidates.Molecules. 2024 Dec 30;30(1):107. doi: 10.3390/molecules30010107. Molecules. 2024. PMID: 39795166 Free PMC article.
-
Synthesis, In Vivo Anticonvulsant Activity Evaluation and In Silico Studies of Some Quinazolin-4(3H)-One Derivatives.Molecules. 2024 Apr 24;29(9):1951. doi: 10.3390/molecules29091951. Molecules. 2024. PMID: 38731442 Free PMC article.
-
Discovery of A Novel Series of Quinazoline-Thiazole Hybrids as Potential Antiproliferative and Anti-Angiogenic Agents.Biomolecules. 2024 Feb 12;14(2):218. doi: 10.3390/biom14020218. Biomolecules. 2024. PMID: 38397456 Free PMC article.
-
Computational Study on the Effect of Inactivating/Activating Mutations on the Inhibition of MEK1 by Trametinib.Int J Mol Sci. 2020 Mar 21;21(6):2167. doi: 10.3390/ijms21062167. Int J Mol Sci. 2020. PMID: 32245216 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

