The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain
- PMID: 25794615
- PMCID: PMC6221458
- DOI: 10.1016/j.molcel.2015.02.015
The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain
Abstract
DNA binding by numerous transcription factors including the p53 tumor suppressor protein constitutes a vital early step in transcriptional activation. While the role of the central core DNA binding domain (DBD) of p53 in site-specific DNA binding has been established, the contribution of the sequence-independent C-terminal domain (CTD) is still not well understood. We investigated the DNA-binding properties of a series of p53 CTD variants using a combination of in vitro biochemical analyses and in vivo binding experiments. Our results provide several unanticipated and interconnected findings. First, the CTD enables DNA binding in a sequence-dependent manner that is drastically altered by either its modification or deletion. Second, dependence on the CTD correlates with the extent to which the p53 binding site deviates from the canonical consensus sequence. Third, the CTD enables stable formation of p53-DNA complexes to divergent binding sites via DNA-induced conformational changes within the DBD itself.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
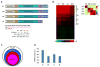


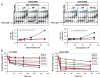


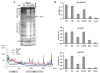
References
-
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. - PubMed
-
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. - PubMed
-
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

