TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation
- PMID: 25794620
- PMCID: PMC4374139
- DOI: 10.1016/j.molcel.2015.02.012
TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation
Abstract
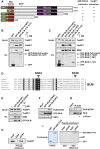
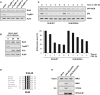
The Bloom syndrome helicase BLM and topoisomerase-IIβ-binding protein 1 (TopBP1) are key regulators of genome stability. It was recently proposed that BLM phosphorylation on Ser338 mediates its interaction with TopBP1, to protect BLM from ubiquitylation and degradation (Wang et al., 2013). Here, we show that the BLM-TopBP1 interaction does not involve Ser338 but instead requires BLM phosphorylation on Ser304. Furthermore, we establish that disrupting this interaction does not markedly affect BLM stability. However, BLM-TopBP1 binding is important for maintaining genome integrity, because in its absence cells display increased sister chromatid exchanges, replication origin firing and chromosomal aberrations. Therefore, the BLM-TopBP1 interaction maintains genome stability not by controlling BLM protein levels, but via another as-yet undetermined mechanism. Finally, we identify critical residues that mediate interactions between TopBP1 and MDC1, and between BLM and TOP3A/RMI1/RMI2. Taken together, our findings provide molecular insights into a key tumor suppressor and genome stability network.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Comment in
-
TopBP1 stabilizes BLM protein to suppress sister chromatid exchange.Mol Cell. 2015 Mar 19;57(6):955-956. doi: 10.1016/j.molcel.2015.02.011. Mol Cell. 2015. PMID: 25794614 No abstract available.
References
-
- Blackford A.N., Schwab R.A., Nieminuszczy J., Deans A.J., West S.C., Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum. Mol. Genet. 2012;21:2005–2016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

