In vivo mapping of the functional regions of the DEAD-box helicase Vasa
- PMID: 25795910
- PMCID: PMC4400588
- DOI: 10.1242/bio.201410579
In vivo mapping of the functional regions of the DEAD-box helicase Vasa
Abstract
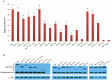
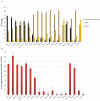
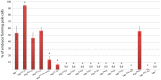
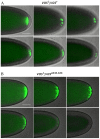
The maternally expressed Drosophila melanogaster DEAD-box helicase Vasa (Vas) is necessary for many cellular and developmental processes, including specification of primordial germ cells (pole cells), posterior patterning of the embryo, piRNA-mediated repression of transposon-encoded mRNAs, translational activation of gurken (grk) mRNA, and completion of oogenesis itself. Vas protein accumulates in the perinuclear nuage in nurse cells soon after their specification, and then at stage 10 Vas translocates to the posterior pole plasm of the oocyte. We produced a series of transgenic constructs encoding eGFP-Vas proteins carrying mutations affecting different regions of the protein, and analyzed in vivo which Vas functions each could support. We identified novel domains in the N- and C-terminal regions of the protein that are essential for localization, transposon repression, posterior patterning, and pole cell specification. One such functional region, the most C-terminal seven amino acids, is specific to Vas orthologues and is thus critical to distinguishing Vas from other closely related DEAD-box helicases. Surprisingly, we also found that many eGFP-Vas proteins carrying mutations that would be expected to abrogate DEAD-box helicase function localized to the nuage and posterior pole, and retained the capacity to support oogenesis, although they did not function in embryonic patterning, pole cell specification, grk activation, or transposon repression. We conclude from these experiments that Vas, a multifunctional protein, uses different domains and different molecular associations to carry out its various cellular and developmental roles.
Keywords: Drosophila; Embryonic patterning; Germ cells; Pole plasm; RNA helicase; piRNA biogenesis.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
Figures



Similar articles
-
C-terminal residues specific to Vasa among DEAD-box helicases are required for its functions in piRNA biogenesis and embryonic patterning.Dev Genes Evol. 2016 Nov;226(6):401-412. doi: 10.1007/s00427-016-0560-5. Epub 2016 Aug 29. Dev Genes Evol. 2016. PMID: 27572922
-
Multiple Functions of the DEAD-Box Helicase Vasa in Drosophila Oogenesis.Results Probl Cell Differ. 2017;63:127-147. doi: 10.1007/978-3-319-60855-6_6. Results Probl Cell Differ. 2017. PMID: 28779316 Review.
-
Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities.Development. 1994 May;120(5):1201-11. doi: 10.1242/dev.120.5.1201. Development. 1994. PMID: 8026330
-
Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation.Curr Biol. 2003 Oct 28;13(21):1905-9. doi: 10.1016/j.cub.2003.10.026. Curr Biol. 2003. PMID: 14588248
-
The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology.Biochim Biophys Acta. 2013 Aug;1829(8):810-6. doi: 10.1016/j.bbagrm.2013.04.005. Epub 2013 Apr 13. Biochim Biophys Acta. 2013. PMID: 23587717 Review.
Cited by
-
Building RNA-protein germ granules: insights from the multifaceted functions of DEAD-box helicase Vasa/Ddx4 in germline development.Cell Mol Life Sci. 2021 Dec 18;79(1):4. doi: 10.1007/s00018-021-04069-1. Cell Mol Life Sci. 2021. PMID: 34921622 Free PMC article. Review.
-
piRNA Biogenesis in Drosophila melanogaster.Trends Genet. 2017 Nov;33(11):882-894. doi: 10.1016/j.tig.2017.09.002. Epub 2017 Sep 27. Trends Genet. 2017. PMID: 28964526 Free PMC article. Review.
-
Germ Cell Lineage Homeostasis in Drosophila Requires the Vasa RNA Helicase.Genetics. 2019 Nov;213(3):911-922. doi: 10.1534/genetics.119.302558. Epub 2019 Sep 4. Genetics. 2019. PMID: 31484689 Free PMC article.
-
The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage.Genes Dev. 2017 May 1;31(9):939-952. doi: 10.1101/gad.297051.117. Epub 2017 May 23. Genes Dev. 2017. PMID: 28536148 Free PMC article.
-
Origin and establishment of the germline in Drosophila melanogaster.Genetics. 2025 Apr 17;229(4):iyae217. doi: 10.1093/genetics/iyae217. Genetics. 2025. PMID: 40180587 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

