SISPA-Seq for rapid whole genome surveys of bacterial isolates
- PMID: 25796360
- PMCID: PMC5556377
- DOI: 10.1016/j.meegid.2015.03.018
SISPA-Seq for rapid whole genome surveys of bacterial isolates
Abstract
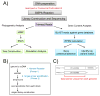
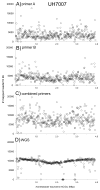
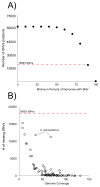
Whole genome sequencing (WGS) of large isolate collections has many applications, yet sequencing costs are still significant. We sought to develop a rapid and cost efficient WGS method to address fundamental questions in clinical microbiology. We evaluated the performance of SISPA (Sequence-Independent, Single-Primer Amplification) combined with next-generation sequencing (SISPA-Seq) of 75 clinical isolates of Acinetobacter baumannii to establish whether SISPA-Seq resulted in sufficient coverage and quality to (1) determine strain phylogenetic placement and (2) and carriage of known antibiotic resistance (AbR) genes. Strains for which whole genome sequences were available were included for validation. Two libraries for each strain were constructed from separate SISPA reactions with different barcoded primers, using genomic DNA prepared from either high quality or rapid heat-lysis preparations. SISPA-Seq resulted in a median of 65× genome coverage when reads from both primer sets were combined. Coverage and quality were sufficient for detection of AbR genes by comparison of reads to the ARG-ANNOT database and were often sufficient to distinguish between different allelic variants of the same gene. kSNP and RAxML were used to construct a robust phylogeny based on single-nucleotide variants (SNVs) that showed that the SISPA-Seq data was sufficient for sensitive and accurate phylogenetic placement. Advantages of the SISPA-Seq method include inexpensive and rapid DNA preparation and a typical total cost less than one-half that of standard genome sequencing. In summary, SISPA-Seq can be used to survey whole genomes of a large strain collection and identify strains that should be targeted for additional sequencing.
Keywords: Acinetobacter baumannii; SISPA; SISPA-Seq; Whole genome survey.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

