Structure and function insights into the NuRD chromatin remodeling complex
- PMID: 25796366
- PMCID: PMC11114056
- DOI: 10.1007/s00018-015-1880-8
Structure and function insights into the NuRD chromatin remodeling complex
Abstract
Transcription regulation through chromatin compaction and decompaction is regulated through various chromatin-remodeling complexes such as nucleosome remodeling and histone deacetylation (NuRD) complex. NuRD is a 1 MDa multi-subunit protein complex which comprises many different subunits, among which histone deacetylases HDAC1/2, ATP-dependent remodeling enzymes CHD3/4, histone chaperones RbAp46/48, CpG-binding proteins MBD2/3, the GATAD2a (p66α) and/or GATAD2b (p66β) and specific DNA-binding proteins MTA1/2/3. Here, we review the currently known crystal and NMR structures of these subunits, the functional data and their relevance for biomedical research considering the implication of NuRD subunits in cancer and various other diseases. The complexity of this macromolecular assembly, and its poorly understood mode of interaction with the nucleosome, the repeating unit of chromatin, illustrate that this complex is a major challenge for structure-function relationship studies which will be tackled best by an integrated biology approach.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


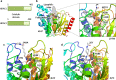

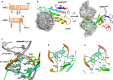

References
-
- Waddington CH. The epigenotype. Int J Epidemiol. 2012;41:10–13. - PubMed
-
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. - PubMed
-
- Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. - PubMed
-
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. - PubMed
-
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

