Bacterial transfer RNAs
- PMID: 25796611
- PMCID: PMC4542688
- DOI: 10.1093/femsre/fuv004
Bacterial transfer RNAs
Abstract
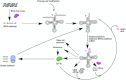
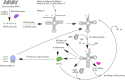
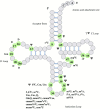
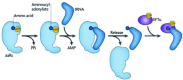
Transfer RNA is an essential adapter molecule that is found across all three domains of life. The primary role of transfer RNA resides in its critical involvement in the accurate translation of messenger RNA codons during protein synthesis and, therefore, ultimately in the determination of cellular gene expression. This review aims to bring together the results of intensive investigations into the synthesis, maturation, modification, aminoacylation, editing and recycling of bacterial transfer RNAs. Codon recognition at the ribosome as well as the ever-increasing number of alternative roles for transfer RNA outside of translation will be discussed in the specific context of bacterial cells.
Keywords: aminoacylation; codon; modification; protein synthesis; tRNA; translation.
© FEMS 2015. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Altman S. Isolation of tyrosine tRNA precursor molecules. Nat New Biol. 1971;229:19–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

