Mechanism of alternative splicing and its regulation
- PMID: 25798239
- PMCID: PMC4360811
- DOI: 10.3892/br.2014.407
Mechanism of alternative splicing and its regulation
Abstract
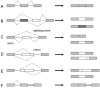
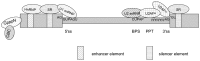
Alternative splicing of precursor mRNA is an essential mechanism to increase the complexity of gene expression, and it plays an important role in cellular differentiation and organism development. Regulation of alternative splicing is a complicated process in which numerous interacting components are at work, including cis-acting elements and trans-acting factors, and is further guided by the functional coupling between transcription and splicing. Additional molecular features, such as chromatin structure, RNA structure and alternative transcription initiation or alternative transcription termination, collaborate with these basic components to generate the protein diversity due to alternative splicing. All these factors contributing to this one fundamental biological process add up to a mechanism that is critical to the proper functioning of cells. Any corruption of the process may lead to disruption of normal cellular function and the eventuality of disease. Cancer is one of those diseases, where alternative splicing may be the basis for the identification of novel diagnostic and prognostic biomarkers, as well as new strategies for therapy. Thus, an in-depth understanding of alternative splicing regulation has the potential not only to elucidate fundamental biological principles, but to provide solutions for various diseases.
Keywords: alternative splicing; disease; mechanism; precursor mRNA; regulation.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
