Analytical methods in untargeted metabolomics: state of the art in 2015
- PMID: 25798438
- PMCID: PMC4350445
- DOI: 10.3389/fbioe.2015.00023
Analytical methods in untargeted metabolomics: state of the art in 2015
Abstract
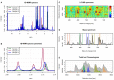
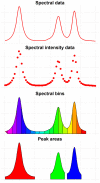
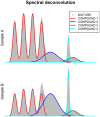
Metabolomics comprises the methods and techniques that are used to measure the small molecule composition of biofluids and tissues, and is actually one of the most rapidly evolving research fields. The determination of the metabolomic profile - the metabolome - has multiple applications in many biological sciences, including the developing of new diagnostic tools in medicine. Recent technological advances in nuclear magnetic resonance and mass spectrometry are significantly improving our capacity to obtain more data from each biological sample. Consequently, there is a need for fast and accurate statistical and bioinformatic tools that can deal with the complexity and volume of the data generated in metabolomic studies. In this review, we provide an update of the most commonly used analytical methods in metabolomics, starting from raw data processing and ending with pathway analysis and biomarker identification. Finally, the integration of metabolomic profiles with molecular data from other high-throughput biotechnologies is also reviewed.
Keywords: data analysis; integration; mass spectrometry; metabolomics; nuclear magnetic resonance; pathway analysis; spectral processing; untargeted.
Figures

References
-
- Akiyama K., Chikayama E., Yuasa H., Shimada Y., Tohge T., Shinozaki K., et al. (2008). PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In silico Biol. 8, 339–345. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

