No evidence of enemy release in pathogen and microbial communities of common wasps (Vespula vulgaris) in their native and introduced range
- PMID: 25798856
- PMCID: PMC4370511
- DOI: 10.1371/journal.pone.0121358
No evidence of enemy release in pathogen and microbial communities of common wasps (Vespula vulgaris) in their native and introduced range
Abstract
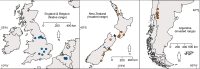
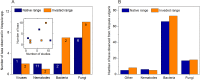
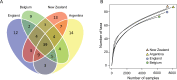
When invasive species move to new environments they typically experience population bottlenecks that limit the probability that pathogens and parasites are also moved. The invasive species may thus be released from biotic interactions that can be a major source of density-dependent mortality, referred to as enemy release. We examined for evidence of enemy release in populations of the common wasp (Vespula vulgaris), which attains high densities and represents a major threat to biodiversity in its invaded range. Mass spectrometry proteomic methods were used to compare the microbial communities in wasp populations in the native (Belgium and England) and invaded range (Argentina and New Zealand). We found no evidence of enemy release, as the number of microbial taxa was similar in both the introduced and native range. However, some evidence of distinctiveness in the microbial communities was observed between countries. The pathogens observed were similar to a variety of taxa observed in honey bees. These taxa included Nosema, Paenibacillus, and Yersina spp. Genomic methods confirmed a diversity of Nosema spp., Actinobacteria, and the Deformed wing and Kashmir bee viruses. We also analysed published records of bacteria, viruses, nematodes and fungi from both V. vulgaris and the related invader V. germanica. Thirty-three different microorganism taxa have been associated with wasps including Kashmir bee virus and entomophagous fungi such as Aspergillus flavus. There was no evidence that the presence or absence of these microorganisms was dependent on region of wasp samples (i.e. their native or invaded range). Given the similarity of the wasp pathogen fauna to that from honey bees, the lack of enemy release in wasp populations is probably related to spill-over or spill-back from bees and other social insects. Social insects appear to form a reservoir of generalist parasites and pathogens, which makes the management of wasp and bee disease difficult.
Conflict of interest statement
Figures



References
-
- Keane RM, Crawley MJ. Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol. 2002;17: 164–170.
-
- Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM. Introduced species and their missing parasites. Nature 2003;421: 628–630. - PubMed
-
- Heger T, Jeschke JM. The enemy release hypothesis as a hierarchy of hypotheses. Oikos 2014;123: 741–750.
-
- Archer ME. A Key to the world species of the Vespinae (Hymenoptera). Res Monogr Coll Ripon St. John 1989;2: 1–41.
-
- Dvorák L. Social wasps (Hymenoptera: Vespidae) trapped with beer in European forest ecosystems. Acta Mus Morav. 2007;92: 181–204.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

