Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes
- PMID: 25798884
- PMCID: PMC4370668
- DOI: 10.1371/journal.pone.0121064
Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes
Abstract
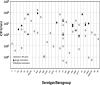
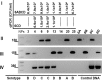
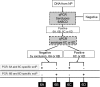
Streptococcus pneumoniae globally kills more children than any other infectious disease every year. A prerequisite for pneumococcal disease and transmission is colonization of the nasopharynx. While the introduction of pneumococcal conjugate vaccines has reduced the burden of pneumococcal disease, understanding the impact of vaccination on nasopharyngeal colonization has been hampered by the lack of sensitive quantitative methods for the detection of >90 known S. pneumoniae serotypes. In this work, we developed 27 new quantitative (q)PCR reactions and optimized 26 for a total of 53 qPCR reactions targeting pneumococcal serotypes or serogroups, including all vaccine types. Reactions proved to be target-specific with a limit of detection of 2 genome equivalents per reaction. Given the number of probes required for these assays and their unknown shelf-life, the stability of cryopreserved reagents was evaluated. Our studies demonstrate that two-year cryopreserved probes had similar limit of detection as freshly-diluted probes. Moreover, efficiency and limit of detection of 1-month cryopreserved, ready-to-use, qPCR reaction mixtures were similar to those of freshly prepared mixtures. Using these reactions, our proof-of-concept studies utilizing nasopharyngeal samples (N=30) collected from young children detected samples containing ≥2 serotypes/serogroups. Samples colonized by multiple serotypes/serogroups always had a serotype that contributes at least 50% of the pneumococcal load. In addition, a molecular approach called S6-q(PCR)2 was developed and proven to individually detect and quantify epidemiologically-important serogroup 6 strains including 6A, 6B, 6C and 6D. This technology will be useful for epidemiological studies, diagnostic platforms and to study the pneumobiome.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

