MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5' end resection
- PMID: 25799990
- PMCID: PMC4481296
- DOI: 10.1038/nature14216
MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5' end resection
Abstract
Appropriate repair of DNA lesions and the inhibition of DNA repair activities at telomeres are crucial to prevent genomic instability. By fuelling the generation of genetic alterations and by compromising cell viability, genomic instability is a driving force in cancer and ageing. Here we identify MAD2L2 (also known as MAD2B or REV7) through functional genetic screening as a novel factor controlling DNA repair activities at mammalian telomeres. We show that MAD2L2 accumulates at uncapped telomeres and promotes non-homologous end-joining (NHEJ)-mediated fusion of deprotected chromosome ends and genomic instability. MAD2L2 depletion causes elongated 3' telomeric overhangs, indicating that MAD2L2 inhibits 5' end resection. End resection blocks NHEJ while committing to homology-directed repair, and is under the control of 53BP1, RIF1 and PTIP. Consistent with MAD2L2 promoting NHEJ-mediated telomere fusion by inhibiting 5' end resection, knockdown of the nucleases CTIP or EXO1 partially restores telomere-driven genomic instability in MAD2L2-depleted cells. Control of DNA repair by MAD2L2 is not limited to telomeres. MAD2L2 also accumulates and inhibits end resection at irradiation-induced DNA double-strand breaks and promotes end-joining of DNA double-strand breaks in several settings, including during immunoglobulin class switch recombination. These activities of MAD2L2 depend on ATM kinase activity, RNF8, RNF168, 53BP1 and RIF1, but not on PTIP, REV1 and REV3, the latter two acting with MAD2L2 in translesion synthesis. Together, our data establish MAD2L2 as a crucial contributor to the control of DNA repair activity by 53BP1 that promotes NHEJ by inhibiting 5' end resection downstream of RIF1.
Figures

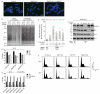
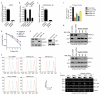


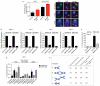



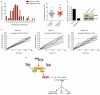
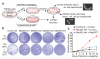
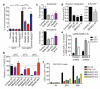


Comment in
-
REV7/MAD2L2: the multitasking maestro emerges as a barrier to recombination.EMBO J. 2015 Jun 12;34(12):1609-11. doi: 10.15252/embj.201591697. Epub 2015 Apr 20. EMBO J. 2015. PMID: 25896508 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. - PubMed
Additional references to Full Methods
-
- Kato H, et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–41. - PubMed
-
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

