Structural imprints in vivo decode RNA regulatory mechanisms
- PMID: 25799993
- PMCID: PMC4376618
- DOI: 10.1038/nature14263
Structural imprints in vivo decode RNA regulatory mechanisms
Erratum in
-
Erratum: Structural imprints in vivo decode RNA regulatory mechanisms.Nature. 2015 Nov 12;527(7577):264. doi: 10.1038/nature15717. Epub 2015 Sep 30. Nature. 2015. PMID: 26416736 Free PMC article. No abstract available.
Abstract
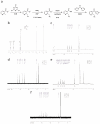
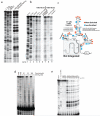
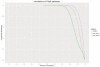
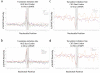
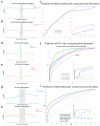
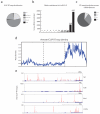
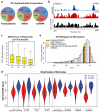
Visualizing the physical basis for molecular behaviour inside living cells is a great challenge for biology. RNAs are central to biological regulation, and the ability of RNA to adopt specific structures intimately controls every step of the gene expression program. However, our understanding of physiological RNA structures is limited; current in vivo RNA structure profiles include only two of the four nucleotides that make up RNA. Here we present a novel biochemical approach, in vivo click selective 2'-hydroxyl acylation and profiling experiment (icSHAPE), which enables the first global view, to our knowledge, of RNA secondary structures in living cells for all four bases. icSHAPE of the mouse embryonic stem cell transcriptome versus purified RNA folded in vitro shows that the structural dynamics of RNA in the cellular environment distinguish different classes of RNAs and regulatory elements. Structural signatures at translational start sites and ribosome pause sites are conserved from in vitro conditions, suggesting that these RNA elements are programmed by sequence. In contrast, focal structural rearrangements in vivo reveal precise interfaces of RNA with RNA-binding proteins or RNA-modification sites that are consistent with atomic-resolution structural data. Such dynamic structural footprints enable accurate prediction of RNA-protein interactions and N(6)-methyladenosine (m(6)A) modification genome wide. These results open the door for structural genomics of RNA in living cells and reveal key physiological structures controlling gene expression.
Figures



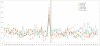


Comment in
-
RNA. Detailed probing of RNA structure in vivo.Nat Rev Genet. 2015 May;16(5):255. doi: 10.1038/nrg3939. Epub 2015 Apr 9. Nat Rev Genet. 2015. PMID: 25854184 No abstract available.
-
Defining Functional Structured RNA inside Living Cells.Biochemistry. 2017 Nov 7;56(44):5847-5848. doi: 10.1021/acs.biochem.7b00816. Biochemistry. 2017. PMID: 29064690 No abstract available.
Similar articles
-
Measuring RNA structure transcriptome-wide with icSHAPE.Methods. 2017 May 1;120:85-90. doi: 10.1016/j.ymeth.2017.02.010. Epub 2017 Mar 20. Methods. 2017. PMID: 28336307 Free PMC article. Review.
-
Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE.Nat Protoc. 2016 Feb;11(2):273-90. doi: 10.1038/nprot.2016.011. Epub 2016 Jan 14. Nat Protoc. 2016. PMID: 26766114 Free PMC article.
-
RNA structure maps across mammalian cellular compartments.Nat Struct Mol Biol. 2019 Apr;26(4):322-330. doi: 10.1038/s41594-019-0200-7. Epub 2019 Mar 18. Nat Struct Mol Biol. 2019. PMID: 30886404 Free PMC article.
-
In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features.Nature. 2014 Jan 30;505(7485):696-700. doi: 10.1038/nature12756. Epub 2013 Nov 24. Nature. 2014. PMID: 24270811
-
RNA binding protein/RNA element interactions and the control of translation.Curr Protein Pept Sci. 2012 Jun;13(4):294-304. doi: 10.2174/138920312801619475. Curr Protein Pept Sci. 2012. PMID: 22708490 Free PMC article. Review.
Cited by
-
A novel end-to-end method to predict RNA secondary structure profile based on bidirectional LSTM and residual neural network.BMC Bioinformatics. 2021 Mar 31;22(1):169. doi: 10.1186/s12859-021-04102-x. BMC Bioinformatics. 2021. PMID: 33789581 Free PMC article.
-
Emerging Perspectives of RNA N6-methyladenosine (m6A) Modification on Immunity and Autoimmune Diseases.Front Immunol. 2021 Mar 5;12:630358. doi: 10.3389/fimmu.2021.630358. eCollection 2021. Front Immunol. 2021. PMID: 33746967 Free PMC article. Review.
-
Long Noncoding RNAs in Cancer Pathways.Cancer Cell. 2016 Apr 11;29(4):452-463. doi: 10.1016/j.ccell.2016.03.010. Cancer Cell. 2016. PMID: 27070700 Free PMC article. Review.
-
Evolving insights into RNA modifications and their functional diversity in the brain.Nat Neurosci. 2016 Sep 27;19(10):1292-8. doi: 10.1038/nn.4378. Nat Neurosci. 2016. PMID: 27669990 Free PMC article.
-
Cognitive neuroepigenetics: the next evolution in our understanding of the molecular mechanisms underlying learning and memory?NPJ Sci Learn. 2016;1:16014. doi: 10.1038/npjscilearn.2016.14. Epub 2016 Jul 20. NPJ Sci Learn. 2016. PMID: 27512601 Free PMC article.
References
-
- Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi:10.1038/nature12756. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

