Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice
- PMID: 25799994
- PMCID: PMC4482136
- DOI: 10.1038/nature14289
Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice
Abstract
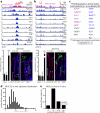
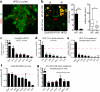
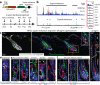
Adult stem cells occur in niches that balance self-renewal with lineage selection and progression during tissue homeostasis. Following injury, culture or transplantation, stem cells outside their niche often display fate flexibility. Here we show that super-enhancers underlie the identity, lineage commitment and plasticity of adult stem cells in vivo. Using hair follicle as a model, we map the global chromatin domains of hair follicle stem cells and their committed progenitors in their native microenvironments. We show that super-enhancers and their dense clusters ('epicentres') of transcription factor binding sites undergo remodelling upon lineage progression. New fate is acquired by decommissioning old and establishing new super-enhancers and/or epicentres, an auto-regulatory process that abates one master regulator subset while enhancing another. We further show that when outside their niche, either in vitro or in wound-repair, hair follicle stem cells dynamically remodel super-enhancers in response to changes in their microenvironment. Intriguingly, some key super-enhancers shift epicentres, enabling their genes to remain active and maintain a transitional state in an ever-changing transcriptional landscape. Finally, we identify SOX9 as a crucial chromatin rheostat of hair follicle stem cell super-enhancers, and provide functional evidence that super-enhancers are dynamic, dense transcription-factor-binding platforms which are acutely sensitive to pioneer master regulators whose levels define not only spatial and temporal features of lineage-status but also stemness, plasticity in transitional states and differentiation.
Figures










References
-
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. - PubMed
-
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 810.1126/science.1196236. Epub 1192010 Sep 1196223. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

