Capturing the dynamics of pathogens with many strains
- PMID: 25800537
- PMCID: PMC4698306
- DOI: 10.1007/s00285-015-0873-4
Capturing the dynamics of pathogens with many strains
Abstract
Pathogens that consist of multiple antigenic variants are a serious public health concern. These infections, which include dengue virus, influenza and malaria, generate substantial morbidity and mortality. However, there are considerable theoretical challenges involved in modelling such infections. As well as describing the interaction between strains that occurs as a result cross-immunity and evolution, models must balance biological realism with mathematical and computational tractability. Here we review different modelling approaches, and suggest a number of biological problems that are potential candidates for study with these methods. We provide a comprehensive outline of the benefits and disadvantages of available frameworks, and describe what biological information is preserved and lost under different modelling assumptions. We also consider the emergence of new disease strains, and discuss how models of pathogens with multiple strains could be developed further in future. This includes extending the flexibility and biological realism of current approaches, as well as interface with data.
Keywords: Cross-immunity; Evolution; Influenza; Multi-strain pathogens; Transmission model.
Figures
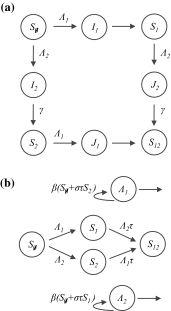

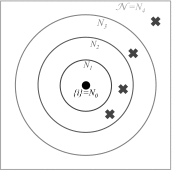

References
-
- Anderson RM, May RM (1991) Infectious Diseases of Humans. Oxford University Press, Dynamics and Control
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

