Global variability in gene expression and alternative splicing is modulated by mitochondrial content
- PMID: 25800673
- PMCID: PMC4417112
- DOI: 10.1101/gr.178426.114
Global variability in gene expression and alternative splicing is modulated by mitochondrial content
Abstract
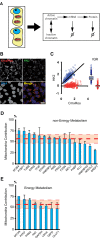
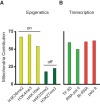
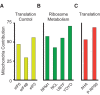
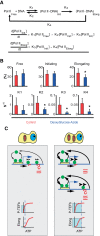
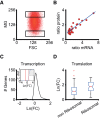
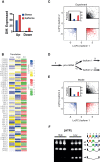
Noise in gene expression is a main determinant of phenotypic variability. Increasing experimental evidence suggests that genome-wide cellular constraints largely contribute to the heterogeneity observed in gene products. It is still unclear, however, which global factors affect gene expression noise and to what extent. Since eukaryotic gene expression is an energy demanding process, differences in the energy budget of each cell could determine gene expression differences. Here, we quantify the contribution of mitochondrial variability (a natural source of ATP variation) to global variability in gene expression. We find that changes in mitochondrial content can account for ∼50% of the variability observed in protein levels. This is the combined result of the effect of mitochondria dosage on transcription and translation apparatus content and activities. Moreover, we find that mitochondrial levels have a large impact on alternative splicing, thus modulating both the abundance and type of mRNAs. A simple mathematical model in which mitochondrial content simultaneously affects transcription rate and splicing site choice can explain the alternative splicing data. The results of this study show that mitochondrial content (and/or probably function) influences mRNA abundance, translation, and alternative splicing, which ultimately affects cellular phenotype.
© 2015 Guantes et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Brock A, Chang H, Huang S. 2009. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 10: 336–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
